Isolation and propagation of human Wharton’s jelly stem cells (hWJSCs)
Human umbilical cords (UC) were obtained with written informed patient consent and approval from the Singapore Ministry of Health, Domain Specific Review Board (DSRB). The hWJSCs were derived from human umbilical cords according to our previously published protocol37,38. Briefly, the umbilical cord was cut into 1–2 cm sections and then dissected lengthwise. Sections were placed with their inner surface faced down into DMEM medium (Invitrogen) containing an enzymatic solution comprised of 2 mg/ml collagenase type I (Invitrogen Life Technologies, Carlsbad, CA), 2 mg/ml collagenase type IV (Invitrogen) and 100 IU of hyaluronidase (Invitrogen) in 100 mm sterile plastic dishes (Becton Dickinson, BD, NJ, USA) and incubated at 37 °C in a 5% CO2-in-air atmosphere for 45 min. Subsequently, the enzymatic solution containing the Wharton’s jelly was transferred to sterile 15 ml tubes (BD), syringed through an 18-gauge needle to release the cells from the jelly before being centrifuged at 300×g for 10 min. The supernatant was then decanted, and the cell pellets were resuspended in an hWJSC culture medium comprised of 80% DMEM high glucose supplemented with 20% fetal bovine serum (FBS), (Biochrom, Berlin, Germany), 1% non-essential amino acids, 2 mM l-glutamine, 0.1 mM β-mercaptoethanol, 1% insulin–transferrin–selenium (ITS), antibiotic–antimycotic mixture (Invitrogen) and 16 ng/ml basic fibroblast growth factor (bFGF) (Millipore Bioscience Research Agents, Temecula, CA).
Experimental setup for simulated lunar microgravity (sµG)
Simulated lunar microgravity
hWJSCs (n = 7) were seeded at a density of 5.0 × 105 cells in T25-tissue culture-treated flasks with vented-caps for the control arm, occluded caps for experimental control and sµG arms. For the control arm, cells were seeded with 5 ml of hWJSC medium and incubated at 37 °C in a 5% CO2-in-air atmosphere for three days (with CO2 gas exchange). For both experimental control and sµG arms, the flasks were completely filled with hWJSC culture medium to avoid air bubbles and shear stress during rotation, which is necessary for sµG. The flasks were tightly sealed with parafilm to prevent any media from leaking and incubated in the same 37 °C incubator as the control arm cells for three days. The sµG flasks were taped onto the rotating platform of a Random Positioning Machine (RPM; Yuri Gravity, Meckenbeuren, Germany), which was placed in the CO2 incubator and run on a pre-programed protocol to stimulate lunar gravity (0.16 G) for 3 days. Each biological replicate (n = 7) was cultured in triplicates that were pooled for analyses.
Post microgravity
After 3 days, hWJSCs (n = 7) were disassociated and seeded at a density of 5 × 105 cells in T25-tissue culture flasks with vented caps for all arms; control, post-experimental control and post-experimental (post- sμG). Across the board, cells were cultured with 5 ml of hWJSC medium and incubated at 37 °C in a 5% CO2-in-air atmosphere for another 3 days (with CO2 gas exchange). Each biological replicate (n = 7) was cultured in triplicates that were pooled for analyses.
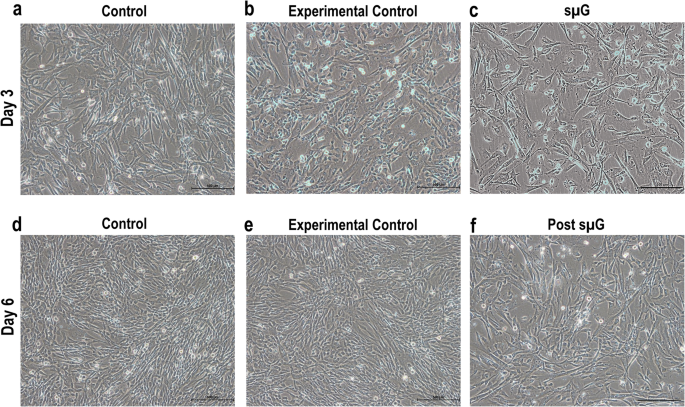
Cell morphology
Cell attachment, morphological changes, and growth of hWJSCs exposed to microgravity and post-microgravity were monitored and photographed using inverted phase contrast optics (Nikon Instruments, Tokyo, Japan).
Trypan blue vital counts
The viability of hWJSCs cultured under sµG and post microgravity were quantified using trypan blue vital counts. An aliquot of each cultured hWJSCs from control, experimental control, and sµG arms at day 3 (sμG) and day 6 (post-sμG) was taken and stained with 0.4% Trypan Blue (vital dye) (Sigma) for 1 min at room temperature. The numbers of live cells (unstained) were either counted manually using a hemocytometer (Hausser Scientific, Horsham, PA, USA) or automatedly using the Luna Automated Cell Counter (Bio Cat, Heidelberg, Germany).
Fluorescence-activated cell sorting (FACS) analysis
hWJSCs were disassociated using TrypLETM Express (Invitrogen) for 3–5 min prior to PBS wash to obtain single-cell suspensions, which were then blocked with 10% normal goat serum (NGS) (Invitrogen Life Technologies, Carlsbad, CA) for 30 min to prevent non-specific binding. The cells were incubated with primary antibodies: CD34, CD45, CD73, CD90, and CD105 (1:100, Biolegend, San Diego, CA) for 1 h followed by incubation with Alexa Fluor®488 (1:500) secondary antibody (Invitrogen Life Technologies, Carlsbad, CA) for 30 min. The cells were washed with PBS and re-suspended in 10% NGS. Finally, the cells were filtered using a 70 µm nylon strainer (BD Bioscience) to remove any cell clumps and then analyzed using a CytoFLEX LX Analyzer (Beckman Coulter, Fullerton, CA).
Annexin V-FITC assay
The annexin V-FITC assay was carried out on hWJSCs from control, experimental control, and sµG exposures to evaluate rates of apoptosis. Briefly, the cells were dissociated with TrypLETM Express (Invitrogen), washed once with phosphate-buffered saline (PBS), and then with Annexin V binding buffer (1×). The cells were stained with 5 μl Annexin V-FITC (Promega) and counterstained with propidium iodide (1 μg/ml) (Promega) for 15 min at room temperature, then analyzed using a CytoFLEX LX Analyzer (Beckman Coulter, Fullerton, CA).
RNA extraction
Total RNA was extracted from hWJSCs cultured under sµG and post-sµG conditions (including control, experimental control, and sµG arms) using the RNeasy Mini kit (Qiagen, Venlo, Netherlands). RNA quality and quantity were measured using a Nanodrop™ Spectrophotometer (Nanodrop Technologies, Wilmington, DW) and an Agilent 2100 Bioanalyzer using an Agilent 6000 Nano RNA kit (both Agilent, Waldbronn, Germany). 1000 ng of total RNA (minimum RIN value 9.0) in a 25 μl volume was submitted for sequencing.
RNA sequencing
RNA-seq library was prepared (3’ directional, polyA enrichment) from total RNA with 150-bp paired-end sequencing on the NovaSeq 6000 system by NovogeneAIT Genomics Singapore. Samples were sequenced to a target depth of 30 million reads.
RNA-seq data processing and quality control
The 150-bp paired-end sequenced reads were processed using the nf-core/rnaseq v3.10.1 pipeline39 with nextflow v22.10.440. Briefly, raw reads were trimmed using Trim Galore! v0.6.741 to remove low-quality bases and adapters. Trimmed reads were aligned to the human reference genome, hg38, with STAR v2.6.1d aligner42. The gene counts were generated using Salmon v1.9.043 quantification of the aligned reads. Principle component analysis was conducted for quality control assessment.
Bioinformatic analysis
All downstream analyses were performed using R statistical software version 4.2.144. Differential gene expression analysis was performed using the DESeq2 package v1.36.045, adjusting for cell line differences. Genes with at least 2-fold change in expression and FDR < 0.05 were considered significant. Results were visualized using the EnhancedVolcano v1.14.046 and ComplexHeatmap v2.12.147. Pathway enrichment analysis (GSEA) was run on all genes ranked by their signed log p-value using clusterProfiler v4.9.048 and KEGG database49. Pathways with FDR < 0.10 were considered significant.
Confirmatory studies
To confirm that the transcriptional changes identified in the initial sµG and post-sμG studies resulted in lineage shifts, we performed an additional set of experiments to induce osteogenic and chondrogenic differentiation in sµG hWJSCs and matched controls. As below, these cells were then assessed for lineage differentiation using quantitative PCR and immunohistochemistry. All osteogenic and chondrogenic differentiation studies were performed in triplicate.
Osteogenic differentiation
For the induction of osteogenic differentiation, hWJSCs from control, experimental control, and sµG were seeded (5.0 × 105 cells/dish) in T25-tissue culture treated flasks and incubated at 37 °C in a 5% CO2-in-air atmosphere for 24 h to allow attachment. The medium was then switched to osteogenic medium containing DMEM supplemented 5% FBS, 0.17 mM l-ascorbic-acid (Sigma, MO), 100 nM dexamethasone (Sigma, MO), antibiotic-antimycotic mixture (Invitrogen) and 10 mM β-glycerophosphate (Sigma, MO) and the cells were cultured for 14 days with fresh changes of medium every 48 h.
Chondrogenic differentiation
For the induction of chondrogenic differentiation, hWJSCs from control, experimental control, and sµG were seeded (5.0 × 105 cells/dish) in T25-tissue culture treated flasks and incubated at 37 °C in a 5% CO2-in-air atmosphere for 24 h to allow attachment. The medium was then changed to chondrogenic medium containing DMEM supplemented with 1% ITS, 0.17 mM l-ascorbic-acid, 100 nM dexamethasone, 1 mM sodium pyruvate, 0.35 mM proline (Sigma, MO), antibiotic–antimycotic mixture (Invitrogen) and 10 ng/ml TGFβ-3 (Sigma, MO) and the cells were cultured for 14 days with fresh changes of medium every 48 h.
Von Kossa staining
To assess hWJSCs’ osteogenic potential, mineralization was evaluated by Von Kossa staining for hWJSCs cultured in an osteogenic medium for 14 days. Briefly, cells were washed with PBS and fixed in a 4% formaldehyde solution for 10 min at room temperature. The cells were then washed with distilled water and stained in 1% silver nitrate solution under UV light for 60 min. The cells were then washed again with distilled water, treated with 3% sodium thiosulfate for 5 min, and counterstained with 1% nuclear fast red for 5 min. The stained cells were washed with distilled water and photographed using inverted phase contrast optics (Nikon Instruments, Tokyo, Japan).
Alcian Blue staining
To evaluate hWJSCs chondrogenic potential, hWJSCs were stained with alcian blue staining for the cells cultured in a chondrogenic medium for 14 days. Briefly, the cells were washed and stained with 0.5% Alcian Blue (Sigma, St. Louis, MO) for 30 min at room temperature and then rinsed with tap water. The slides were counterstained with 0.1% Nuclear Fast Red (Sigma, St. Louis, MO) for 5 min and then photographed using inverted phase contrast optics (Nikon Instruments, Tokyo, Japan).
Quantitative real-time polymerase chain reaction (qRT-PCR)
RNA samples were transcribed to cDNA using Tetro cDNA Synthesis kit (Bioline, Eveleigh NSW, Australia). qRT-PCR analysis was performed using ABI PRISM 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA) using SYBR green master mix (Applied Biosystems). The final primer concentration in each reaction per well was 1 μM, and 20 ng of cDNA was used for each reaction well of 96-well plate. The cycling conditions were as follows: initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the normalization control, and untreated respective samples were used as the calibrator. The relative quantification was performed using the comparative CT (2−ΔΔCT) method. The results were expressed as mean ± SD from three replicates for individual experiments.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- PlatoHealth. Biotech and Clinical Trials Intelligence. Access Here.
- Source: https://www.nature.com/articles/s41526-024-00397-1
