Cell culture
All cell lines used and derived by different approaches in this study are listed in Supplementary Table 1. Detailed information about the experimental design, materials and reagents is presented in the Reporting Summary. Primary human adult dermal fibroblasts (HDFa) from three different female donors were obtained from Gibco (C-013-5C, lot no. 1029000 for 38F and lot no. 1569390 for 32F) and cultured following the manufacturer’s recommendations. In brief, cells were thawed and plated into flasks in Medium 106 (Gibco) supplemented with low serum growth supplement (LSGS) (Gibco) for expansion. Cells were cultured in a 37 °C, 5% O2 and 5% CO2 incubator, and the medium was changed every other day. The use of human embryonic stem cells (H9 and MEL1) was carried out in accordance with approvals from Monash University and the Commonwealth Scientific and Industrial Research Organisation (CSIRO) Human Research Ethics Offices. Conventional primed-hiPS cells and H9 hES cells (WiCell Research Institute; http://www.wicell.org) were maintained as described in the below section. The cell lines used in this study were regularly tested and were mycoplasma negative. Human dermal fibroblasts and NHEKs were authenticated by ThermoFisher and Lonza, respectively, as per description in the CoA. hES cells were authenticated in the Laslett lab. MSCs were authenticated in the Heng lab. These cell lines were also routinely authenticated in-house via morphological assessment, immunofluorescence for identity markers, or RNA-seq.
Cell culture media
Fibroblast medium: DMEM (ThermoFisher), 10% fetal bovine serum (FBS, Hyclone), 1% non-essential amino acids (ThermoFisher), 1 mM GlutaMAX (ThermoFisher), 1% penicillin-streptomycin (ThermoFisher), 55 μM β-mercaptoethanol (ThermoFisher) and 1 mM sodium pyruvate (ThermoFisher). Naive medium (t2iLGoY)19: 50:50 mixture of DMEM/F12 (ThermoFisher) and neurobasal medium (ThermoFisher), supplemented with 2 mM l-glutamine (ThermoFisher), 0.1 mM β-mercaptoethanol (ThermoFisher), 0.5% N2 supplement (ThermoFisher), 1% B27 supplement (ThermoFisher), 1% penicillin-streptomycin (ThermoFisher), 10 ng ml−1 human leukaemia inhibitory factor (made in-house), 250 μM l-ascorbic acid (Sigma), 10 μg ml−1 recombinant human insulin (Sigma), 1 μM PD0325901 (Miltenyi Biotec), 1 μM CHIR99021 (Miltenyi Biotec), 2.5 μM Gö6983 (Tocris), 10 μM Y-27632 (Abcam). Primed hiPS cell medium (KSR/FGF2): DMEM/F12 (ThermoFisher), 20% knockout serum replacement (KSR) (ThermoFisher), 1 mM GlutaMAX (ThermoFisher), 0.1 mM β-mercaptoethanol (ThermoFisher), 1% non-essential amino acids (ThermoFisher), 50 ng ml−1 recombinant human FGF2 (Miltenyi Biotec), 1% penicillin-streptomycin (ThermoFisher). Primed hiPS cell medium (Essential 8 (E8)): 10 ml of E8 supplement (Gibco) to 500 ml medium basal (Gibco), supplemented with 1% penicillin-streptomycin (Gibco).
Derivation of TNT-hiPS cells and NTP-hiPS cells
Human somatic cell reprogramming was performed as previously described16,22,48. In brief, early passages (<P6) fibroblast cells were seeded into 6-well plates at 50,000–70,000 cells per well before transduction in fibroblast medium. Cells in one well were trypsinized for counting to determine the volume of virus required for transduction (multiplicity of infection), and transduction was performed using the CytoTune 2.0 iPSC Sendai Reprogramming Kit (Invitrogen) consisting of four transcription factors (OCT4, SOX2, MYC and KLF4). Twenty-four hours later, the medium was removed, with subsequent medium changes performed every other day. For the derivation of primed-hiPS cells, cells were reseeded onto a layer of iMEFs on day 7 of reprogramming and transitioned to primed medium (KSR/FGF2 or E8 on vitronectin; Supplementary Table 1) on the next day. The cells were cultured to confluency (around day 18–21 of reprogramming) and further passaged with Collagenase IV (ThermoFisher) for cell line establishment. For derivation of TNT-hiPS cells, the day 7 reprogramming intermediates were transitioned to naive medium (t2iLGoY) instead. When dome-shaped colonies were evident 5 days later, intermediate cells were collected using Accutase (Stem Cell Technologies) and reseeded onto a layer of iMEFs in naive conditions. The medium was switched to primed medium (KSR/FGF2 or E8; Supplementary Table 1) the following day. When the culture became confluent, cells were collected using collagenase IV and maintained in primed medium (KSR/FGF2 or E8; Supplementary Table 1) on iMEFs. Cells were cultured in a 37 °C, 5% O2 and 5% CO2 incubator with daily medium change. Cells are usually passaged every 4–5 days. For derivation of NTP-hiPS cells: after 16–18 days post-transduction (8–10 days in naive condition), naive-hiPS cells were collected using Accutase (Stem Cell Technologies) and passaged more than 10 times. The established naive-hiPS cells were confirmed by flow cytometry and immunostaining for naive pluripotency-associated markers. Naive-hiPS cells were then collected using Accutase (Stem Cell Technologies) and reseeded in naive condition, the medium was then switched to Primed hiPSC medium (E8) the following day. When the culture became confluent, cells were collected using Collagenase IV (ThermoFisher) and maintained in Primed hiPSC medium (E8). Cells were cultured in 37 °C, 5% O2 and 5% CO2. All cell lines were tested by CGH array and reported normal.
Estimations of cell diversity by Cas9 enrichment for lentivirus insertion mapping
To prepare enriched Oxford Nanopore Technologies (ONT) sequencing libraries, we used PoreChop to design 2 guide RNAs (gRNAs) (5′-AGATCCGTTCACTAATCGAATGG-3′ and 5′-GGAACAGTACGAACGCGCCGAGG-3′) for Cas9-mediated cleavage approximately 1 kb within each end of the integrated lentiviral sequences. These gRNAs were designed to not match elsewhere in the hg38 human reference genome. We confirmed their on-target efficiency by Cas9 (IDT: Alt-R S.p. Cas9 Nuclease V3; catalogue no. 1081058) cleavage of the lentiviral DNA, visualized on gel, in a separate experiment. DNA dephosphorylation (NEB: Quick CIP; M0525S), single guide (IDT: Alt-R CRISPR–Cas9 CRISPR RNA (crRNA) and Alt-R CRISPR–Cas9 trans-activating crRNA (tracrRNA); catalogue no. 1072532) and RNP formation, Cas9 cleavage and subsequent library preparation (ONT: SQK-CS9109) were largely performed according to the ONT Cas9 enrichment guidelines. We increased the starting amount of DNA to 5 µg, and the dephosphorylation and cleavage incubation times to 2 h and 24 h, respectively. For two replicates of each reprogramming method, we then loaded 350 ng of the enriched DNA library onto a MinION R9.4 flow cell, as per the manufacturer’s recommendations, and sequenced for 48 h. Additionally, for the 32F fibroblast sample, 3 µg of unenriched DNA was sequenced on a PromethION R9.4 flow cell (library prep kit SQK-LSK110) by the Kinghorn Centre for Clinical Genomics (KCCG). For data analysis, reads with a Phred score ≥10 were basecalled with Guppy (version 5.0.11). These reads were mapped with minimap2 (version 2.17) to both the human reference genome (hg38), and the sequence of the expected lentiviral insert49. Alignment maps were filtered with samtools (version 1.13) to only keep primary alignments with a length ≥800 bp, and a mapping quality50 of 60. Reads that mapped to both hg38 and the lentivirus sequence were retained and then subjected to another round of filtering. Here, reads were discarded when the base pair interval between the alignments to the lentiviral sequence and hg38 on the read was ≥51 bp. Reads that originated from the unenriched library and comprised a complete (≥4,500 bp) putative lentiviral insert, spanned by a genomic alignment, as identified by TLDR (version 1.2.2) were kept51. Exact insert sites per read were identified based on the coordinates of both alignment maps (hg38 and lentiviral) to the original read. Exact insert sites were clustered together with bedtools (version 2.30.0) cluster within a 50-bp interval52. For each cluster, the coverage was calculated and the smallest start and largest end coordinates were selected as the exact insert site.
The diversity of cell populations was estimated by a Poisson bootstrap53. Here, we model a Poisson distribution of total insertion landscape based on the sequencing coverage of unique lentiviral insert sites. This model infers the amount of non-sequenced insertion sites, which in return is used to adapt the model until convergence, and results in an estimate for the lentiviral insertion diversity.
Secondary fibroblast reprogramming system
hES cells were cultured in fibroblast medium without FGF2 containing DMEM, 10% FBS, 1 mM l-glutamine, 100 µM MEM non-essential amino acids, and 0.1 mM β-mercaptoethanol, for a week. Cells were passaged three times using 0.25% trypsin and then sorted for THY1+TRA160− populations.
Neural stem cell differentiations
hiPS cells were cultivated in E8 medium (Life Technologies) on Cultrex (R&D Systems) coated TC dishes and split 1∶10 every 5 days. Colonies were mechanically disaggregated with 0.5 mM EDTA in PBS (Sigma). After splitting, pieces of colonies were collected by sedimentation and resuspended in E8 medium with 10 μM ROCK inhibitor (Selleckchem) and cultured in petri dishes to form embryoid bodies in suspension. After 24 h, the medium was changed to Knockout DMEM (Life Technologies) with 20% Knockout Serum Replacement (Life Technologies), 1 mM β-mercaptoethanol (Sigma), 1% non-essential amino acids (NEAA, Life Technologies), 1% penicillin/streptomycin (Life Technologies) and 1% Glutamax (Life Technologies) supplemented with 10 µM SB-431542 (Selleckchem), 1 µM dorsomorphin (Selleckchem) for neural induction, as well as 3 µM CHIR99021 (Cayman Chemical) and 0.5 µM PMA (Sigma). Medium was replaced on day 3 by N2B27 medium (50% DMEM-F12 (Life Technologies), 50% Neurobasal (Life Technologies) with 1∶200 N2 supplement (R&D Systems), 1∶100 B27 supplement lacking vitamin A (Miltenyi Biotec) with 1% penicillin-streptomycin (Life Technologies) and 1% Glutamax (Life Technologies)) supplemented with the same small molecule supplements. On day 4, SB-431542 and dorsomorphin were withdrawn and 150 µM ascorbic acid (Sigma) was added to the medium. On day 6, the embryoid bodies were triturated with a 1,000 µl pipette into smaller pieces and plated on Cultrex-coated 12-well plates at a density of about 10–15 per well in NSC expansion medium (N2B27 with CHIR, PMA, and ascorbic acid). After another 5 days, cells were split at a ratio of 1:5 using Trypsin-EDTA (Life Technologies) and Trypsin inhibitor (Sigma) onto a new Cultrex-coated well. After another 5 days, cells were collected by 10 min trypsinization at 37 °C to generate a single-cell suspension for scRNA-seq workflow.
Endoderm progenitor differentiation
The endoderm differentiation was adapted and performed as previously described54,55. In brief, hiPS cells were collected and replated onto plates coated with Matrigel and cultured in primed hiPS cell medium (KSR/FGF2) with medium change for an additional day before differentiation. To differentiate into endodermal progenitor cells, the cells were cultured in chemically defined medium containing 100 ng ml−1 activin A, 20 ng ml−1 FGF2, 10 ng ml−1 bone morphogenetic factor 4 (BMP4), and 10 µM LY294002 for 3–4 days and assessed for differentiation efficiency.
Cortical neuron differentiation
hiPS cells were seeded onto flasks coated with Matrigel at a density of 0.5–1 × 104 cells per cm2 in primed hiPS cell medium (KSR/FGF2). After 48 h, the medium was changed to neural induction medium containing DMEM/F12, B27 without vitamin A supplement (Gibco, ThermoFisher Scientific), N2 supplement (Gibco, ThermoFisher Scientific), 0.1% β-mercaptoethanol (Gibco, ThermoFisher Scientific), 0.66% bovine serum albumin (Sigma-Aldrich), 1% sodium pyruvate (Gibco, ThermoFisher Scientific), 1% non-essential amino acids (Gibco, ThermoFisher Scientific), 1% penicillin and streptomycin, 100 ng ml−1 LDN193189 (Tocris Bioscience, Bio-Techne) for 14 days.
Skeletal muscle cell differentiation
hiPS cells were seeded onto flasks coated with Matrigel at a density of 0.5–1 × 104 cells per cm2 in primed hiPS cell medium (KSR/FGF2). After 24 h, medium was changed to DMEM/F12-based medium supplemented with ITS (insulin + transferrin + selenium; Sigma-Aldrich) with 1% penicillin and streptomycin (Gibco, ThermoFisher Scientific), 3 µM CHIR99021 (Miltenyi Biotec), 0.5 µM LDN193189 (Tocris Bioscience, Bio-Techne) for 3 days. On days 4–6, the medium was changed to DMEM/F12-based medium supplemented with ITS and 3 µM CHIR99021, 20 ng ml−1 FGF2 (Miltenyi Biotec), 0.5 µM LDN193189. On days 7–8, the medium was changed to DMEM/F12-based medium supplemented with 20 ng ml−1 FGF2, 0.5 µM LDN193189, 2 ng ml−1 IGF1 (Peprotech). On days 9–30, the medium was changed to DMEM/F12-based medium supplemented with 15% knockout serum replacement (Gibco, ThermoFisher Scientific), 1% penicillin and streptomycin, 0.05 mg ml−1 BSA (Sigma-Aldrich), 2 ng ml−1 IGF1.
Lung alveolar type 2 cell differentiation
Induced pluripotent stem cells were seeded onto flasks coated with Matrigel at a density of 0.5–1 × 104 cells per cm2 in primed hiPS cell medium (KSR/FGF2). After 48 h, the medium was changed daily with RPMI-based medium with B27 supplement (Gibco, ThermoFisher Scientific), 100 ng ml−1 activin A (Peprotech), 1 µM CHIR99021, 1% penicillin and streptomycin for 3 days. On days 4–8, the medium was changed daily with DMEM/F12-based medium with N2 (Gibco, ThermoFisher Scientific) and B27 supplements, 0.05 mg ml−1 ascorbic acid (Sigma-Aldrich), 0.4 mM monothioglycerol (Sigma-Aldrich), 2 µM dorsomorphin (Peprotech), 10 µM SB-431542 (Miltenyi Biotec), 1% penicillin and streptomycin. On days 9–12, the medium was changed daily with DMEM/F12-based medium with B27 supplement, 0.05 mg ml−1 ascorbic acid, 0.4 mM monothioglycerol, 20 ng ml−1 BMP4 (Peprotech), 0.5 µM all-trans retinoic acid (Sigma-Aldrich), 3 µM CHIR99021, 1% penicillin and streptomycin. On days 12–20, the medium was changed every other day with DMEM/F12-based medium with B27 supplement, 0.05 mg ml−1 ascorbic acid, 0.4 mM monothioglycerol, 10 ng ml−1 FGF10 (Stemcell Technologies), 10 ng ml−1 FGF7 (Peprotech), 3 µM CHIR99021, 50 nM dexamethasone (Sigma-Aldrich), 0.1 mM 8-bromoadenosine 3′,5′-cyclic monophosphate (Sigma-Aldrich), 0.1 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich), 1% penicillin and streptomycin.
Flow cytometry
To obtain a single-cell suspension for flow cytometric analysis or sorting experiments, cells were collected using TrypLE express (Life Technologies) and resuspended in labelling mix (PBS, 2% FBS, 10 µM ROCK inhibitor Y-27632). Reprogramming intermediates and mature hiPS cells were labelled in a stepwise manner for cell surface markers. Step 1: F11R (mouse IgG antibody; 1:150), SSEA3-PE (rat IgM antibody; 1:10, BD Biosciences); step 2: Alexa Fluor 647 goat anti-mouse IgG (1:2,000, ThermoFisher), PE anti-rat IgM (1:200 eBioscience); step 3: CD13-PE-Cy7 (1:400, BD Biosciences), BV421-EpCAM (1:100, BD), TRA-1-60-BUV395 (1:100, BD Biosciences). Cells were incubated for 10 min on ice and then washed with PBS and resuspended in FACS buffer (PBS, 2% FBS, 10 µM Y-27632 and PI (1 in 500)). Prior to sorting, cells were passed through a 35-μm nylon filter. Sorted cells were collected for replating or downstream analyses. For differentiation experiments, cultures were dissociated using Accutase (Stemcell Technologies) and pelleted at 400g for 5 min. For neural differentiation experiments, cells were then resuspended in APC CD57 antibody (322314; Biolegend) and BUV395 CD56 antibody (563554; BD Biosciences); for muscle differentiation experiments, cells were resuspended in PE-Cy7 CD146 antibody (562135; BD Biosciences), BUV395 CD56 antibody (563554; BD Biosciences); for lung differentiation experiments, cells were resuspended in BV421 CD47 antibody (323116; Biolegend) and Brilliant Violet 421 CD326 antibody (324220; Biolegend); for NSC differentiation experiments, cells were labelled with BUV395 CD56 (NCAM) antibody and Alexa647 FAP antibody (FAB3715R; R&D Systems). Cells were resuspended in 2% fetal bovine serum (FBS; Gibco, ThermoFisher Scientific) and PBS (Gibco, ThermoFisher Scientific) and incubated for 15 min at 4 °C. The cell suspension was washed with PBS and pelleted at 400g for 5 min for analysis. Viability of cells was determined using propidium iodide solution (P4864; Sigma-Aldrich). Samples were analysed using an LSR IIb analyser (BD Biosciences) or a FACSAria II cell sorter (BD Biosciences) using BD FACSDiva software (BD Biosciences).
Immunostaining
Cells were fixed in 4% PFA (Sigma), permeabilized with 0.5% Triton X-100 (Sigma) in DPBS (ThermoFisher), and blocked with 5% goat serum (ThermoFisher). All antibodies used in this study are detailed in Supplementary Table 9 (for example, primary antibodies used were rabbit anti-NANOG polyclonal (1:100, Abcam) and mouse anti-TRA-1-60 IgM (1:300, BD Biosciences)). Primary antibody incubation was conducted overnight at 4 °C on shakers followed by incubation with secondary antibodies (1:400) for 1 h. After labelling, cells were stained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI) (1:1,000, ThermoFisher) for 30 min. Images were taken using an IX71 inverted fluorescent microscope (Olympus). The following markers were assessed for respective differentiation assays: SOX17 and FOXA2 for endoderm progenitor differentiation experiments; SOX1 and PAX6 for neural differentiation experiments; PAX3 and PAX7 for skeletal muscle differentiation experiments; GATA6 and TTF1 for lung differentiation experiments.
Quantitative PCR with reverse transcription
RNA was extracted from cells using RNeasy micro kit (Qiagen) or RNeasy mini kit (Qiagen) and QIAcube (Qiagen) according to the manufacturer’s instructions. Reverse transcription was then performed using QuantiTect reverse transcription kit (Qiagen). Real-time PCR reactions were set up in duplicate using QuantiFast SYBR Green PCR Kit (Qiagen) and then carried out on the 7500 Real-Time PCR system (ThermoFisher) using LightCycler 480 software. The GAPDH gene was used to calculate the relative expression of each assessed gene. Information regarding the PCR primers used in this study is available in Supplementary Table 9.
WGBS library preparation
Genomic DNA was isolated with the Qiagen Blood and Tissue Kit according to the manufacturer’s instructions. 0.5% (w/w) of unmethylated lambda phage DNA (Promega) was added to the sample genomic DNA for the purpose of an unmethylated control to measure the bisulfite non-conversion frequency in each sample. Genomic DNA was fragmented with either either a Covaris S2 sonicator or a Covaris M220 sonicator to a mean length of 200 bp, then end-repaired, A-tailed, ligated to methylated Nextflex Bisulfite-Seq barcodes (Perkin Elmer) using the NxSeq AmpFREE low DNA library kit (Gene Target Solutions) and subjected to PCR amplification with KAPA HiFi Uracil+ DNA polymerase (KAPA Biosystems)56. Sequencing was performed single-end on a HiSeq 1500, NextSeq 500, or paired-end on a NovaSeq 6000 (Illumina).
polyA RNA-seq
RNA was extracted using the Agencourt RNAdvance Cell v2 (Beckman Coulter) system following the manufacturer’s instruction with one additional DNAse (NEB) treatment step. RNA amounts and RINe scores were assessed on a TapeStation using RNA Screen Tape (Agilent), and 500 ng of total RNA were used per sample to generate RNA-seq libraries. ERCC ExFold RNA Spike-In mixes (Thermo Scientific) were added as internal control. Libraries were prepared using the TruSeq Stranded mRNA library prep kit (Illumina), using TruSeq RNA unique dual index adapters (Illumina). Libraries were quantified by qPCR on a CFX96/C1000 cycler (Bio-Rad) and sequenced on a NovaSeq 6000 (Illumina) in 2× 53-bp paired-end format.
ATAC–seq
Approximately 106 freshly collected cells were pelleted and washed in PBS, then resuspended in 1 ml of RSB buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40, 0.1% Tween-20, 0.01% Digitonin). After 10 min incubation on ice, samples were spun at 500g for 5 min and resuspended in 500 µl RSB without NP-40 or digitonin, then strained through a 30-µm filter and pelleted again. Resulting nuclei were counted using trypan blue and 50,000 nuclei were resuspended in 25 µl of 2× TD buffer (20 mM Tris-HCl, 10 mM MgCl2, 20% dimethyl formamide). Tagmentation mix was completed by adding 100 U of loaded Tn5, 16.5 µl PBS, 0.5 µl of 1% digitonin and 0.5 µl of Tween-20 to a final volume of 50 µl, followed by incubation for 30 min at 37 °C with 1,000 rpm mixing on a thermo block. After tagmentation, samples were cleaned up using the Qiagen MinElute PCR purification kit. Eluate was amplified using NEBNext 2× MasterMix and Nextera-based adapters as primers. After 10 PCR cycles, a double-sided bead purification was performed using 0.5× and 1.8× Ampure XP beads. Libraries were quantified by qPCR on a CFX96/C1000 cycler (Bio-Rad) and sequenced on a NovaSeq 6000 (Illumina) in 2× 61-bp paired-end format.
H3K9me3 ChIP–seq
Cells were crosslinked for 10 min in 1% formaldehyde and quenched in 125 mM glycine. Prior to ChIP, antibodies were bound to beads by mixing 3 µg H3K9me3 antibody (Abcam, ab8898) with 50 µl washed Dynabead M-280 Sheep Anti-Rabbit IgG (ThermoFisher) in 500 µl RIPA-150 buffer (50 mM Tris-HCl pH 8.0, 0.15 M NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100 and 0.1% sodium deoxycholate) and incubated at 4 °C for 6 h on a rotator. Crosslinked cells were lysed on ice for 10 min in 15 ml ChIP lysis buffer (50 mM HEPES pH 7.9, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) supplemented with 1x EDTA-free Protease Inhibitor Cocktail (Roche). Lysed cells were centrifuged at 3,200g for 5 min, supernatant removed and followed by two washes with 10ml ChIP wash buffer (10 mM Tris-Cl pH 8.0, 200 mM NaCl and 1 mM EDTA pH 8.0). Lysed cells were resuspended in 130 µl nuclei lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA and 1% SDS) supplemented with 1× EDTA-free Protease Inhibitor Cocktail (Roche), transferred to Covaris tubes (microTUBE AFA Fiber 6 × 16 mm) and sheared with the Covaris (S220) for 5 min (5% duty cycle, 200 cycles per burst and 140 watts peak output at 4 °C). Sheared chromatin was transferred to 1.5 ml eppendorf tubes, centrifuged at 10,000g for 10 min. The supernatant was transferred to 2 ml low-bind tubes containing 1.2 ml ChIP dilution Buffer (50 mM Tris-HCl pH 8.0, 0.167 M NaCl, 1.1% Triton X-100 and 0.11% sodium deoxycholate) and 0.65 ml RIPA-150 buffer, and incubated with the previously prepared H3K9me3 antibody bound Dynabeads at 4 °C overnight on a rotator. Chromatin bound beads were subsequently washed one time with 1 ml RIPA-150 buffer, two times with 1 ml RIPA-500 buffer (50 mM Tris-HCl pH 8.0, 0.5 M NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100 and 0.1% sodium deoxycholate), two times with 1ml RIPA-LiCl buffer (50 mM Tris-HCl pH 8.0, 1 mM EDTA, 1% NP-40, 0.7% sodium deoxycholate and 0.5 M LiCl2) and two times with TE buffer (10 mM Tris-HCl, pH 8.0, 0.1 mM EDTA). After wash steps, DNA was eluted, crosslinks were reversed, and immunoprecipitated DNA was purified by Agencourt AMPure XP beads (Beckman Coulter, A63880). Libraries were prepared from ChIP eluate containing 10 ng DNA using the SMARTer ThruPLEX DNA-Seq Kit (Takara) with SMARTer DNA unique dual index (Takara). After limited PCR amplification, libraries were purified using Agencourt AMPure XP beads (Beckman Coulter), and eluted in a final volume of 20 µl. Libraries were sequenced on a NovaSeq 6000 (Illumina).
scRNA-seq
Single-cell suspensions were counted using a haemocytometer and 200,000 cells per sample used for incubation with hashtag antibodies. Cells were filtered through a 40 µm cell strainer, centrifuged at 800g for 5 min and resuspended in a total volume of 46 µl cell staining buffer (2% BSA (Sigma), 0.01% Tween (Sigma) in 1× DPBS (Life Technologies)) with 4 µl of Fc blocking reagent (Biolegend) and incubated for 10 min on ice. Then, each sample received 0.2 µg of a different TotalSeq-A anti-human Hashtag antibody (Biolegend) and was incubated for 30 min on ice for antibody binding. After the incubation, 1 ml of cell staining buffer was added, and sample centrifuged at 300g for 3 min. Supernatant was removed and cells washed again for a total of three washes to remove all unbound antibodies. Cells were counted, and equal cell numbers for each sample combined to get a cell concentration suitable for loading on the 10x Chromium controller aiming to get 10,000 cells represented. The mixed cell suspension was filtered one more time using a 40-µm cell strainer and processed for scRNA-seq using the 10x Genomics 3′ v3 chemistry following the manufacturer’s instructions. Libraries for scRNA-seq were made following the standard workflow, while HTO libraries for hashtag information were generated as follows: during the cDNA amplification step, HTO primers were added to allow amplification of the HTO barcodes, and supernatant from the first step of clean-up after cDNA amplification PCR was not discarded but used to prepare the HTO library. HTO products were purified using 2x SPRI beads and amplified for 8 PCR cycles with 10× SI-PCR oligo and TruSeq Small RNA RPIx primers to generate a library of ~180 bp fragment size. Sequencing was performed on a NovaSeq 6000 to generate ~420 million reads for the scRNA-seq library and ~40 million reads for the HTO library.
WGBS methylation analysis
Sequencing adapters were trimmed with BBduk with the options mink = 3, qtrim = r, trimq = 10 minlength = 20 before alignment to hg19 with Bowtie and BSseeker2 with the option -n 157,58. PCR duplicates were removed using Sambamba59 and DNA methylation levels at base resolution calculated using CGmap tools60. The non-conversion rate was calculated using the DNA methylation levels for the spiked–in lambda phage genome. When DNA methylation levels were calculated for regions such as promoters, enhancers, DMRs or ICRs, DNA methylation levels were calculated as a coverage-weighted mean by summing the number of methylated C calls (mC) and dividing that by the total number of reads with either a C or T call (C), for the CG or CA dinucleotide contexts separately (defined as mCG/CG and mCA/CA, respectively). To calculate methylation in CH contexts (where H is A, T or C), the level of methylation was calculated as above (mCH/CH) with the non-conversion rate subtracted from this value. When CH methylation was calculated for individual contexts, for example CA methylation, the non-conversion rate for that context was subtracted from the calculated methylation levels. For CA methylation browser tracks, mCA/CA was calculated for 5 kb sliding windows (1-kb slide), with the CA methylation non-conversion rate for that library subtracted from each window. To calculate per-read methylation, reads classified as methylated had methylation calls at every CG position in the read; unmethylated reads had zero methylation calls at CG positions; partially methylated reads had at least one CG methylation call and one non-methylated CG call.
DMR analyses
To test for differentially methylated regions between hiPS cells and hES cells, we first collapsed the stranded mCG values to obtain one value for the symmetrical CG dinucleotides and then performed DMR testing using DMRseq with the options bpSpan = 500, maxGap = 500, maxPerms = 10 and subsequently filtered for DMRs61 with mCG/CG difference >0.2 and P value < 0.05. For CH-DMR analyses, we used the CH-DMRs as previously defined13. We took each CH-DMR and equivalent upstream and downstream genomic regions and divided them into 30 equal-length bins and calculated mCA/CA for each bin and then flank-normalized the binned mCA/CA values by dividing them by their maximum value.
Quantification of gene and transposable element expression
PolyA RNA-seq (Fig. 4 and Extended Data Fig. 10): adapters were trimmed using fastp with default parameters62, and mapped to hg19 using HISAT2 with the options–no-mixed–dta–rna-strandness RF -k 263. Alignments were then filtered to keep only unique mapping read pairs using Samtools view -F “[NH]==1”50. Gene and transposable element read counts were calculated using TEtranscripts and the TElocal script and the curated TE GTF files for hg19 that accompany this software64. Differential expression testing was performed using the glmLRT function within edgeR and genes were determined as significant if log2FC was <1, FDR <0.05 and average log counts per million for the gene was >1. When testing for differential expression of individual transposable elements, we obtained a matrix that contained counts for all genes and individual transposable elements, then filtered this for low or not expressed elements using the filterByExpr function and then calculated the normalization factors for the count matrix. We then performed differential expression testing on this matrix using the glmLRT function to obtain fold-change and significance values. As we were not testing for differential expression of genes, but wanted to retain their counts for library normalization, we then filtered the fold-change and significance table to only include the transposable elements, and then recalculated the FDR for transposable elements only. Significant transposable elements were then classed as differentially expressed if log2FC was <1, FDR <0.05 and average log2 counts per million for the transposable element was >0.
ATAC–seq analysis
Sequencing adapters were trimmed with BBduk with the options mink = 3, ktrim = r, before alignment to hg19 with Bowtie2 with the option -X 2000. Reads were filtered for proper pairs, and PCR duplicates and mitochondrial reads removed using SAMtools. Bigwig browser tracks were normalized for library size using the counts per million method at single base resolution. ATAC–seq peaks were called with MACS2 with the options–nomodel–keep-dup all–gsize hs. Reads counts in peaks for each library were calculated using the summarizeOverlaps function in the GenomicAlignments R package. Differential peak analyses were performed using EdgeR with the glmQLFit glmQLFTest functions. ATAC–seq peaks were considered differentially expressed if the FDR was <0.05, the average log counts per million was >1, and the absolute log2FC was >2. Although we observed differences in ATAC–seq peak counts for NTP-hiPS cells that were not consistent with DNA methylation or gene expression for two outlier samples (Fig. 4b–d), we believe this is due to an additional freeze-thaw cycle for the ATAC–seq samples, and the extended recovery of these two replicates which required two additional passages.
H3K9me3 ChIP–seq analysis
Adapters were trimmed using fastp with default parameters62, and mapped to hg19 using bowtie2 with the option -X 2000. H3K9me3 fold enrichment was calculated for each ChIP and associated input library using the MACS2 bdgcmp function with the option -FE. H3K9me3 fold-enrichment values and peaks for primary fibroblasts and hES cells were downloaded from the ENCODE database for the following accessions: ENCFF735TXC (fibroblast H3K9me3 fold enrichment bigwig file); ENCFF963GBQ (fibroblast H3K9me3 peaks); ENCFF108MOZ (hES cell H3K9me3 fold enrichment bigwig); ENCFF001SUW (hES cell H3K9me3 peaks).
Regulatory element principal component analysis, c-means clustering and motif enrichment analysis
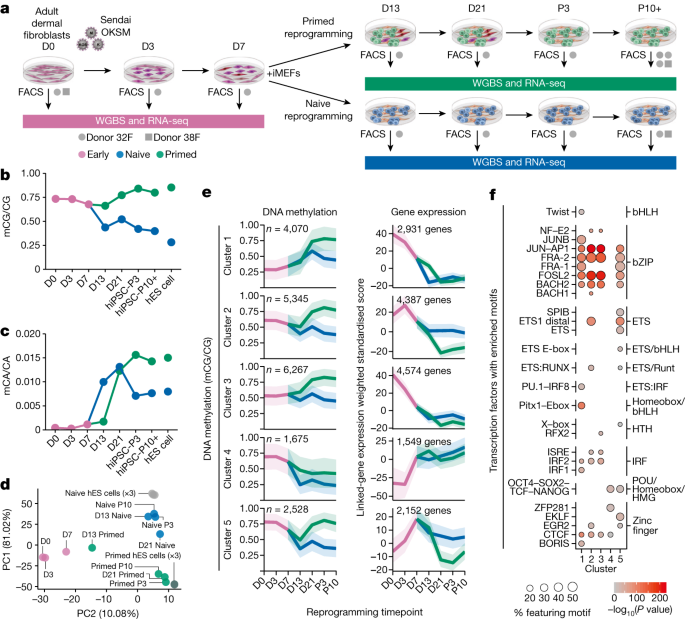
DNA methylation levels were calculated for GeneHancer promoter and enhancer elements using the ‘ClusteredInteractionsDoubleElite’ elements47 in the UCSC hg19 table browser. These regulatory elements include a linked gene and a confidence score for gene linkage. For principal component analysis (PCA) and c-means clustering (Fig. 1d), we calculated the coverage-weighted mean methylation level (mCG/CG) for all the regulatory elements. Principal components were calculated using the R function pr. For Fig. 1e, c-means clustering was performed on regulatory elements that featured ≥20% mCG change at any time through primed reprogramming. Clusters were then identified for both the primed and naive reprogramming time courses with the functions included with the R package Mfuzz65, highly overlapping clusters between the two time courses merged. To plot the expression of genes for each cluster, we first calculated the transcripts per million (TPM) for all genes and then quantile-normalized the gene-expression matrix. Each gene-expression measure was then weighted by enhancer interaction score (TPM × interaction score) to down-weight the expression of linked genes with low interaction scores as many elements were linked to more than one gene. The gene-expression plots in Fig. 1e shows the mean weighted and normalized gene-expression value and the 99% confidence interval. Gene ontology was performed on cluster genes using g:Profiler66. Enriched motifs for each cluster were identified using HOMER with findMotifsGenome.pl and the options hg19 -size given67.
Genomic feature enrichment analysis
To perform association analysis of genomic regions we performed permutation tests calculate enrichment of genomic elements with elements obtained from the GeneHancer database47; ultra-conserved elements as defined previously68; repeat elements as defined by UCSC repeat masker for hg19; fibroblast partially methylated domains calculated for day_0 fibroblasts with MethylSeeker69; promoters defined as 2 kb upstream and 500 bases downstream of TSS as defined in UCSC genes; Exons and introns as defined in UCSC genes; LADs for fibroblasts (4DNFIUIDLJJI) and H1 ES cells (4DNFIP6N54B3) as defined by 4D nucleome project for hg38 and lifted over to hg19 coordinates70,71. H3K9me3 peaks were retrieved from the ENCODE database for fibroblasts (ENCFF963GBQ) and hES cells (ENCFF001SUW)72. Constitutive regions for LADs or H3K9me3 were defined as those regions where peaks intersected for both fibroblasts and hES cells. In these enrichment analyses, the permutation tests calculate how many overlaps the features of interest (that is, CG-DMRs) have, for example, with fibroblast-specific H3K9me3 regions compared to randomly selected regions, and permuted 200 times. This approach addresses the problem of simply comparing the percentage of overlaps, as one does not know how many of those occur by chance. The z-scores from the permutation testing are a measure of the strength of the association, and is defined as the distance between the expected value and the observed one, measured in standard deviations. For example, a z-score of +25 would indicate that the number of overlaps is 25 standard deviations higher than one would expect by chance.
Gene ontology
All gene ontology analyses were performed using g:Profiler using default options and the background set as all detectable genes in the dataset being tested66.
scRNA-seq analysis
RNA-seq fastq files were processed using CellRanger count 3.1.0, while HTO fastq files were processed using CITE-seq-Count 1.4.3 using parameters -cbf 1 -cbl 16 -umif 17 -umil 26 -cells 10000 and feeding sequences of oligonucleotide barcodes. RNA and HTO data were loaded into Seurat 3.1.1 and combined by intersecting cell barcodes found in both datasets. RNA data was log normalized, variable features detected by mean variance while HTO data was normalized by centred log-ratio transformation with margin = 1. Mitochondria were removed based on low UMI counts and enrichment for mitochondrial transcripts. HTODemux was used with positive.quantile = 0.99 to assign single cells back to their sample origins and to exclude doublets and negatives from further analysis. Top 1000 most variable features were used for scaling and PCA of RNA data, using 10 dimensions with a resolution of 0.6 for clustering and UMAP. Cluster identities were defined based on the expression of markers for mesoderm (BMP1, BMP4, HAND1, SNAI1, TGFB1 and TGFB2), endoderm (AFP, ALB, CLDN6, FABP1, FOXA1 and HNF4A) and neural stem cells (NCAM1, NES, NR2F1, PAX3, SOX1 and SOX2). No clusters expressing markers of pluripotency (FUT4, KLF4, MYC, NANOG, POU5F1 and ZFP42) could be detected. By using the HTO identity for each singlet cell, the proportion of cell identities within each of the samples used could be defined.
Statistics and reproducibility
The experiments on characterizing the cell lines derived in this study were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment. All the experiments have been performed as at least two independent experiments as indicated in Methods or figure legends. The derivation of respective primed and TNT-iPS cells has been performed in four biological replicates (four cell types: primary HDFs, NHEK cells, MSCs and our hES cell-derived secondary fibroblast isogenic reprogramming system (secondary fibroblasts) as described in this Article) and was repeated in three independent reprogramming experiments. For the differentiation assays performed in Fig. 5 and Extended Data Fig. 10, a summary of the sample size can be found in Supplementary Table 10.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Automotive / EVs, Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- PlatoHealth. Biotech and Clinical Trials Intelligence. Access Here.
- ChartPrime. Elevate your Trading Game with ChartPrime. Access Here.
- BlockOffsets. Modernizing Environmental Offset Ownership. Access Here.
- Source: https://www.nature.com/articles/s41586-023-06424-7
