Cell culture
Commercially available hiPSC lines were used in this study (Supplementary Table 1). HiPSC lines were obtained from RIKEN Cell Bank (201B7, 253G1, 409B2, HiPS-RIKEN-1A, HiPS-RIKEN-2A, and HiPS-RIKEN-12A), American Type Culture Collection (ATCC-DYR0110 hiPSC and ATCC-HYR01103 hiPSC), JCRB Cell Bank (Tic), and System Biosciences (human mc-iPS). HiPSCs were screened for mycoplasma contamination and hiPSCs used in this study were mycoplasma-free. Undifferentiated hiPSCs were maintained on an iMatrix-511 (Nippi) in StemFit AK02 medium (Ajinomoto). All cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2 and 95% air.
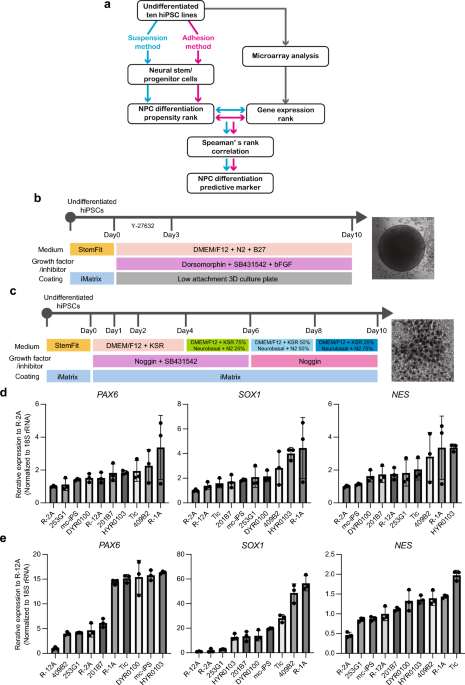
Neural stem/progenitor cell differentiation of hiPSCs
Differentiation of hiPSCs into NS/PCs was induced, as previously reported, with a few modifications. For adhesive differentiation, hiPSCs were detached through incubation with StemPro Accutase (Thermo Fisher Scientific) containing 10 μM Y-27632 for 10 min and seeded onto 24-well cell culture plates (BD Biosciences) coated with iMatrix at a density of 25,000 cells/cm2 for 2–3 days before NS/PC induction. Confluent hiPSCs were treated with 10 μM of the ALK inhibitor SB431542 (Stemgent) and 500 ng/mL of Noggin (R&D systems) in DMEM/F12 medium containing 20% KSR. The medium was replaced on days 1 and 2. On day 6 of differentiation, SB431542 was withdrawn, and increasing amounts of N2 media (25%, 50%, and 75%) were added to the knockout serum replacement medium every 2 days while maintaining 500 ng/mL of Noggin. For suspension differentiation, hiPSCs were treated with 10 μM Y-27632 for 1 h at 37 °C and dissociated with StemPro Accutase (Thermo Fisher Scientific) containing 10 μM Y-27632 for 10 min to generate single-cell suspensions and suspended in B27N2-based medium [DMEM/F12 with 15 mM HEPES, 5% B27, and 5% N2 supplements (Life Technologies), 10 μM SB431542, 2 μM Dorsomorphin (Fujifilm), and 10 ng/mL bFGF (R&D systems)]. The completely dissociated cells were seeded into ultralow attachment 96-well plates (PrimeSurface® 96-well, Sumitomo Bakelite) at 9,000 cells/well, centrifuged at 700 g for 3 min (quick aggregation). The medium was changed daily for up to 10 days; for the first 3 days, 10 µM of Y-27632 was added. Total RNA was obtained from 40 wells of neuro spheres per sample. For microarray analysis, hiPSCs were differentiated into NS/PCs using a STEMdiff SMADi Neural Induction Kit (Stem Cell Technologies) according to the manufacturer’s instructions. Briefly, hiPSCs were maintained on an iMatrix-coated plate in StemFitAK02 media (Ajinomoto) before NS/PC induction. Cells were harvested using Accutase (Thermo Fisher Scientific); 2 × 106 cells were transferred to a Matrigel-coated 6-well plate in STEMdiff Neural Induction Medium + SMADi (Stem Cell Technologies) supplemented with 10 μM Y-27632. The medium was replenished daily with warmed (37 °C) STEMdiff Neural Induction Medium + SMADi until the culture was terminated. Cells were passaged every 7 days, and RNA was extracted from cells harvested at passages (days 7, 14, and 21).
qRT-PCR and PCA
Total RNA was isolated from hiPSCs or differentiated cells using the RNeasy Mini Kit (Qiagen) and treated with DNase I according to the manufacturer’s instructions. qRT-PCR was performed using a QuantiTect Probe One-Step RT-PCR Kit (Qiagen) on a STEPONEPLUS Real-Time PCR System (Applied Biosystems). The expression levels of target genes were normalized to those of the GAPDH transcript or 18S rRNA, which were quantified using TaqMan human Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control reagents (Applied Biosystems) or eukaryotic 18S rRNA endogenous controls (Applied Biosystems), respectively. The probes and primers were obtained from Sigma-Aldrich. The used primer and probe sequences are listed in Supplementary Table 2. PCA was performed using SYSTAT 13 software (Systat Software Inc.) after data standardization (z-scoring) for each NS/PC marker gene.
Rank correlation analysis
To identify microarray probe sets related to the differentiation of hiPSCs into NS/PC, correlations between the intensity value rank of the filtered probe sets and the PC1 rank in the 10 hiPSC lines were determined by calculating Spearman’s rank correlation coefficients (rs), as described in a previous study26. Probe sets exhibiting statistically significant correlations (P < 0.01) were selected. When n = 10 data points, the observed value of rs should exceed 0.794 (positively correlated) or less than –0.794 (negatively correlated) to be considered statistically significant (P < 0.01).
Lentivirus-derived RNAi and generation of ROR2 KD cell lines
ROR2 KD cells were generated by infecting R-2A cells with MISSION Lentiviral Transduction Particle expressing ROR2-targeted shRNAs (#1: TRCN0000199888, #2: TRCN0000001492) or MISSION®pLKO.1-puro Control Non-Mammalian shRNA Control Transduction Articles (Sigma, SHC002V), according to the manufacturer’s instructions. Media containing viruses were collected 48 h after transfection, and the cells were transduced with the viruses using 8 µg/mL polybrene (Sigma-Aldrich) for 24 h. The cells were selected using 2 µg/mL puromycin (Gibco) for 48 h.
Western blotting analysis
The cell lysates were used for western blotting analysis. Proteins were separated using sodium dodecyl sulfate–polyacrylamide gel electrophoresis, transferred to PVDF membranes (Bio-Rad), and blocked for 60 min in Blocking One (Nacalai tesque). Primary antibody dilutions were prepared in Can Get Signal immunoreaction enhancer solution (TOYOBO) as follows: anti-ROR2 antibody (AF2064; R&D Systems) 1:1000, anti-β-actin antibody (A5441; Sigma-Aldrich) 1:2000. Membranes were incubated with HRP-conjugated anti-mouse IgG (Invitrogen) or anti-goat IgG (Invitrogen). Proteins were visualized using ECL Prime Western Blotting Detection Reagent (GE Healthcare) and the ChemiDoc Touch Imaging System (Bio-Rad).
Immunofluorescence staining
HiPSC-derived NS/PC or forebrain neuron was fixed in 4% paraformaldehyde in PBS (Nacalai) for 20 min at 25 °C. After washing with PBS, the cells were permeabilized with 0.2% Triton-X100 (Merk) in PBS for 15 min and blocked with Blocking One (Nacalai) for 30 min. The samples were incubated for 1 h with primary antibodies (anti-PAX6 antibody [PRB-278P-100, BioLegend], anti-MAP2 antibody [MAB8304, R&D systems], and anti-GAD1 antibody [AF2086, BioLegend]). Indirect immunostaining was performed with the secondary antibody (anti-rabbit IgG/Alexa Fluor 555 [A27039, Thermo Fisher Scientific], anti-goat IgG/Alexa Fluor 488 [A32814, Thermo Fisher Scientific], and anti-mouse IgG/Alexa Fluor 488 [A28175, Thermo Fisher Scientific]) for 1 h and examined under a BZ-X810 fluorescence microscope (Keyence).
Generation of a ROR2-overexpressing cell line
ROR2 overexpression cells were generated by infecting 253G1 cells with lentiviral particles expressing ROR2. Briefly, the nucleotide sequence of the human ROR2 open reading frame (NM_004560) was de novo synthesized (Eurofins Genomics) and cloned into the pLVSIN-EF1α puromycin vector (Takara Clontech). Lentivirus packaging and virus infection were performed as described above.
Microarray (Clariom D Assay)
Total RNA was extracted from hiPSC-derived NS/PC cells using an RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. Total RNA (100 ng per sample) was used as the input for the Clariom D Assay (Thermo Fisher Scientific). Target preparation was performed using a Gene Chip™ WT PLUS Reagent Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Hybridization was performed in a Gene Chip Hybridization Oven 645 for 16 h at 45 °C. Gene chips were scanned using a GeneChip Scanner 3000. Array quality control was performed using Transcriptome Analysis Console software (version 4.0.2.15). The National Center for Biotechnology Information Gene Expression Omnibus (NCBI GEO) accession number for the microarray data is GSE233228.
Mature neuron differentiation
Differentiation of hiPSCs into mature nerves was performed according to the manufacturer’s instructions using the STEMdiff Forebrain Neuron Differentiation Kit (#08600, STEMCELL Technologies) for forebrain-type nerves and the STEMdiff Midbrain Neuron Differentiation Kit (#100-0038, STEMCELL Technologies) for midbrain nerves. Using the STEMdiff SMADi Neural Induction Kit (Stem Cell Technologies) monolayer culture protocol described above, hiPSCs were differentiated into NS/PC, and mature neural differentiation was induced.
For midbrain neuron differentiation, hiPSC-derived NS/PCs (day 21, passage 3) were detached using Accutase and seeded into PLO (Sigma)-and laminin (Sigma)-coated 12-well plate at a density of 1.25 × 105 cells/cm2 culture in STEMdiff Neural Induction Medium + SMADi medium for 24 h. The complete medium was replaced daily for 6 days with STEMdiff Midbrain Neuron Differentiation Medium. The midbrain neural precursors (day 7) were detached using ACCUTASE and seeded into PLO-and Laminin-coated 12-well plate at a density of 5 × 104 cells/cm2 in STEMdiff Midbrain Neuron Maturation medium with a half-medium change every 2–3 days for 14 days.
For forebrain-type neuron differentiation, hiPSC-derived NS/PCs (day 21, passage 3) were detached using Accutase and then seeded into PLO-and Laminin-coated 12-well plate at a density of 1.25 × 105 cells/cm2 culture in STEMdiff Neural Induction Medium + SMADi medium for 24 h. The full medium was replaced daily for 6 days with STEMdiff Forebrain Neuron Differentiation medium. The forebrain neural precursors (day 7) were detached using Accutase and seeded into PLO- and Laminin-coated 12-well plate at a density of 5 × 104 cells/cm2 in STEMdiff Forebrain Neuron Maturation media with a half-medium change every 2–3 days for 14 days.
Statistical analysis
Statistical analyses were performed using Prism 9 software (version 9.5.1; GraphPad Software Inc.). Data are presented as mean ± standard deviation (SD). For comparison between two groups the t-test was applied; in cases where another statistic test was applied, it is mentioned accordingly. Statistical significance was set at P < 0.05.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- PlatoHealth. Biotech and Clinical Trials Intelligence. Access Here.
- Source: https://www.nature.com/articles/s41598-023-51082-4
