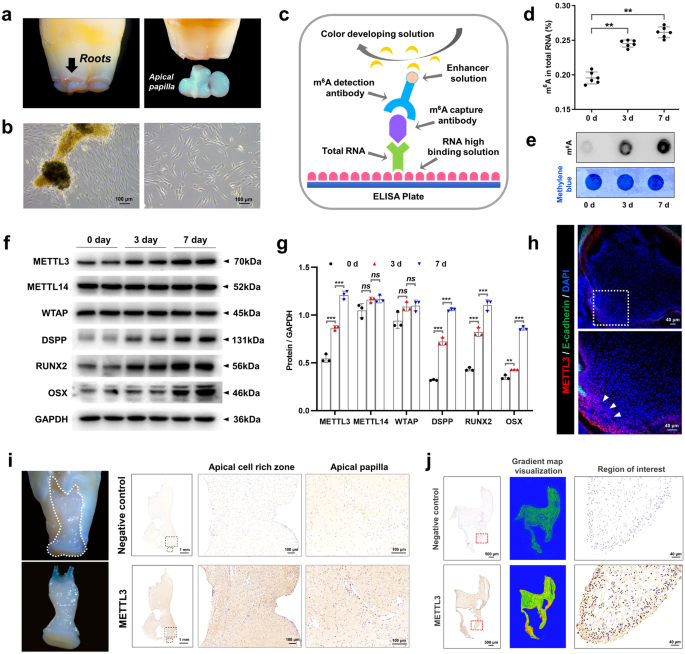
METTL3 expression is positively correlated with odonto/osteoblast differentiation in SCAPs
SCAPs were isolated and exhibited a characteristic spindle-like morphology (Fig. 1a, b). Flow cytometric analysis confirmed that the extracted SCAPs were undifferentiated stem cells derived from the mesenchyme, as these cells expressed CD29, CD73, CD90, and CD105 but did not express CD34 or CD45 (Supplementary Fig. 1). Additionally, the ability of SCAPs to differentiate into three lineages, namely, chondrogenic, osteogenic, and adipogenic lineages, was confirmed through Alcian blue staining, Alizarin red staining, and Oil red O staining, respectively (Supplementary Fig. 2). The results indicate that SCAPs have the ability to differentiate in multiple directions.
a, b SCAPs were isolated according to the literature. SCAPs displayed a typical spindle-like morphology in the original and third generations. Scale bar = 100 µm. c, d Annotation of the m6A RNA methylation quantification kit and the expression of m6A at 0, 3, and 7 days of mineralization; n = 6. e m6A dot blot showing an increase in global m6A levels during osteogenic induction in SCAPs at 3 and 7 days. n = 3. f, g Western blotting was performed to analyze the expression of METTL3, METTL14 and WTAP during SCAP odonto/osteogenic differentiation at 0, 3 and 7 days. n = 3. h Immunofluorescence staining was conducted on CD1 mice at postnatal Day 7. Scale bar = 40 µm. i Representative images of immunohistochemistry assays demonstrating the enrichment of METTL3 in the papilla tissue of human apical teeth. PBS was used as a negative control. Scale bar = 1 m; 100 µm. j Immunohistochemically stained samples of the METTL3-enriched zone in the papilla tissue of human apical teeth. PBS was used as a negative control. Scale bar = 500 µm; 40 µm. ns indicates no significance; ** indicates P < 0.01; *** indicates P < 0.001.
To assess METTL3-mediated m6A modification during odontogenic induction, we cultured SCAPs in odontogenic induction medium for 0, 3, or 7 days. The results demonstrated a gradual increase in the overall level of m6A during SCAP mineralization (Fig. 1c–e). Additionally, western blotting analysis revealed a progressive increase in METTL3 expression with prolonged induction time, accompanied by increased expression of odonto/osteoblast markers (DSPP/RUNX2/OSX). Conversely, the expression of the main “writer” proteins WTAP and METTL14 did not significantly differ (Fig. 1f, g). To observe the in vivo expression pattern of METTL3, we conducted immunofluorescence staining on postnatal Day 7 CD1 mouse samples. The findings demonstrated that METTL3 is primarily present in the apical papilla tissue of tooth roots (Fig. 1h). Additionally, immunohistochemistry assays confirmed that enriched localization was observed in the human dental papilla (Fig. 1i). METTL3 was also expressed in the apical tissue of third molars that displayed almost complete root development (Fig. 1j). In general, these findings indicate that METTL3-induced m6A alterations might affect the differentiation of odonto/osteoblasts in SCAPs.
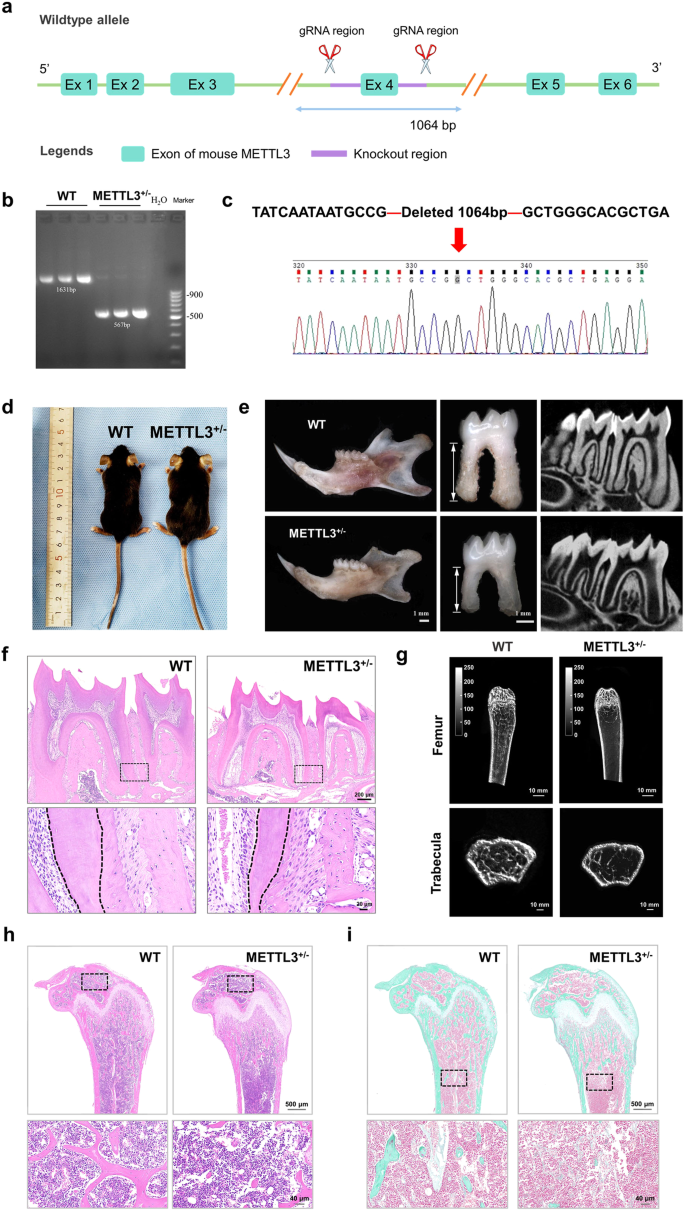
METTL3 knockdown (METTL3+/−) mice exhibit tooth root and bone dysplasia
To investigate the role of METTL3 in tooth root and bone development, we established a METTL3 knockdown mouse model using the CRISPR/Cas9 system. As METTL3 homozygous knockout (METTL3-/-) mice showed early embryonic lethality, we used METTL3 heterozygous knockdown (METTL3+/−) mice for subsequent experiments22. The genotype was initially confirmed through PCR and sequencing and further verified by western blotting (Fig. 2a–d, Supplementary Fig. 3a). H&E staining revealed no discernible differences in the size or morphology of the lung, spleen, kidney, liver, or heart between METTL3+/− mice and wild-type (WT) mice (Supplementary Fig. 3b). Subsequently, stereoscopic microscopy and micro-CT analyses of mandibular first molars revealed a reduction in molar root length and root dentin width in METTL3+/− mice compared to their WT littermates. Furthermore, the mandibular first molars of METTL3+/− mice exhibited a significant reduction in dentin width, as evidenced by H&E staining (Fig. 2e, f). Additionally, the results obtained from micro-CT, H&E staining, and Goldner staining collectively indicated that METTL3 knockdown significantly decreased bone mass and bone formation (Fig. 2g-i). Taken together, the above results suggest the important and notable role of METTL3 in root development and bone formation.
a–d Schematic illustration of the generation of METTL3+/− knockout mice (C57BL/6 N) via a CRISPR/Cas9-mediated genome engineering strategy. Six exons were identified. Exon 4 was selected as the target site. Cas9 and guide RNA (gRNA) were coinjected into fertilized eggs for mouse production, followed by genotype identification of METTL3+/− KO mice. e A reduction in root length and dysplasia of the tooth root in METTL3+/− knockout mice compared to WT mice were observed via stereoscopic microscopy and micro-CT analyses. Scale bar = 1 mm. f Representative images of HE staining indicating that the width of the root dentin in the mandibular first molar was greater in METTL3 +/- mice than in WT mice. Scale bar = 200 µm; 20 µm. g Representative micro-CT images of the femoral metaphysis and entire proximal femur. Scale bar = 10 mm. h, i Representative images of HE staining and Golder staining showing that METTL3+/− mice exhibited reduced bone formation relative to that of their WT counterparts. Scale bar = 500 µm; 40 µm.
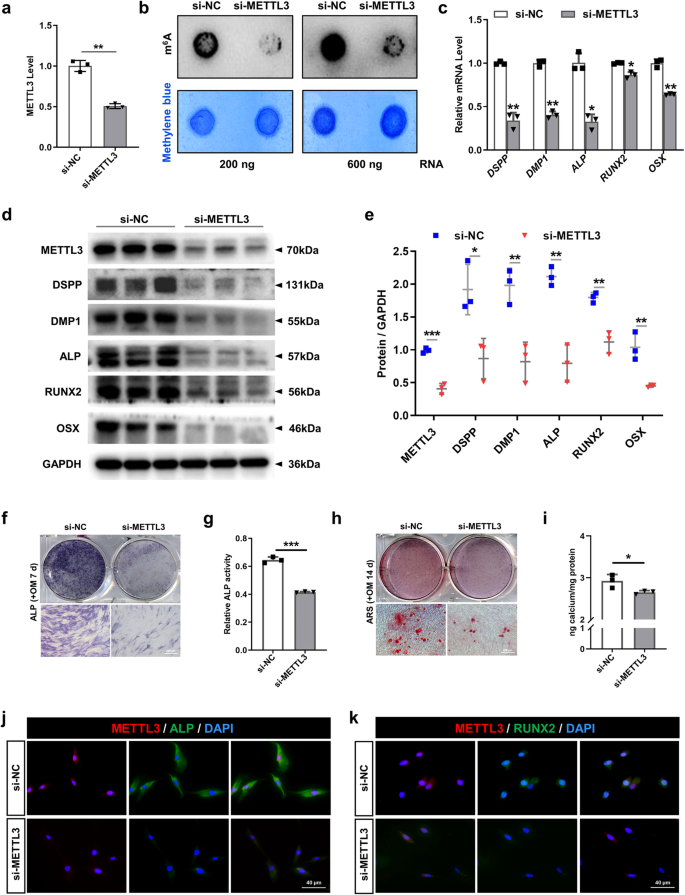
METTL3 positively regulates the odonto/osteogenic differentiation capacity of SCAPs
To assess the regulatory impact of METTL3 on the odontogenic/osteogenic differentiation of SCAPs, we suppressed METTL3 via transfection of specific small interfering RNAs (siRNAs) (Fig. 3a). As expected, m6A dot blot analysis revealed a decrease in the overall m6A level in SCAPs upon METTL3 knockdown (Fig. 3b). Furthermore, the expression levels of genes and proteins associated with odonto/osteogenic differentiation decreased, whereas METTL14 did not significantly change (Fig. 3c–e, Supplementary Fig. 3c). Reduced ALP staining and activity, as well as decreased ARS staining, revealed a diminished degree of odonto/osteogenesis and mineralization in SCAPs (Fig. 3f–i). Immunofluorescence staining analysis additionally demonstrated a significant reduction in the protein expression levels of RUNX2 and ALP in the SCAPs transfected with si-METTL3 compared to those in the si-NC group (Fig. 3j, k).
a The efficiency was measured by qRT‒PCR in SCAPs transfected with si-METTL3. b m6A dot blot assay showing the negative effects of METTL3 inhibition on the m6A content. c qRT‒PCR showed that the mRNA levels of DSPP, DMP1, ALP, RUNX2 and OSX were decreased upon METTL3 silencing. d, e The protein expression of METTL3, DSPP, DMP1, ALP, RUNX2 and OSX decreased after METTL3 inhibition. f–i Representative images of ALP staining and ARS staining and relative quantification showing the negative effects of METTL3 silencing compared with si-NC treatment at 7 and 14 days in the mineralized medium of SCAPs. Scale bar = 200 µm. j, k Representative images of immunofluorescence staining for METTL3, ALP and RUNX2 in SCAPs after METTL3 silencing. Scale bar = 40 µm. n = 3, * indicates P < 0.05; ** indicates P < 0.01; *** indicates P < 0.001.
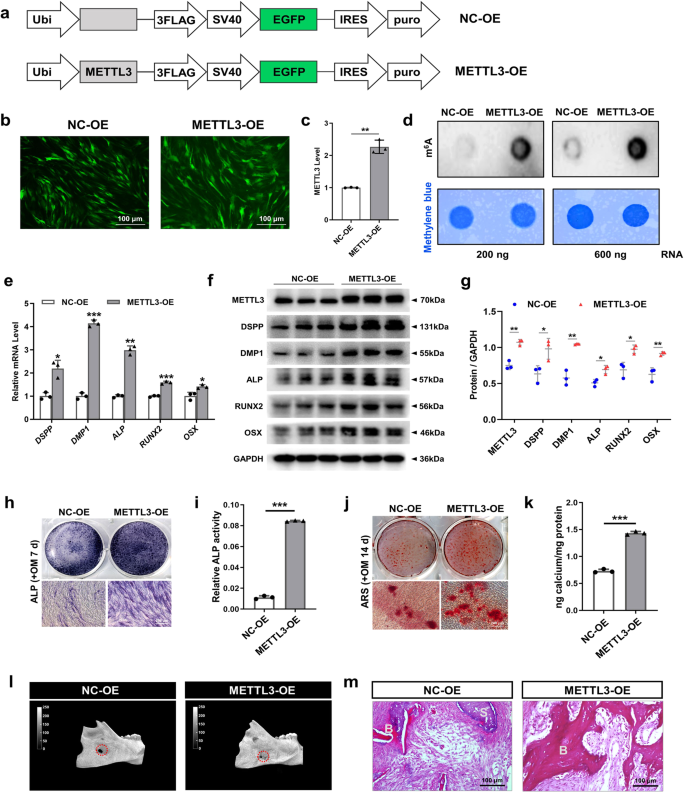
In contrast, the impact of METTL3 overexpression on the differentiation of SCAPs was investigated using recombinant lentiviruses to introduce METTL3 (METTL3-OE) into SCAPs (Fig. 4a–c). Overexpression of METTL3 led to elevated levels of m6A, as shown by the m6A dot blot assay (Fig. 4d). qRT‒PCR and western blotting revealed that the upregulation of METTL3 substantially increased the levels of odonto/osteogenic biomarkers (Fig. 4e‒g). ALP staining and ARS staining revealed a greater number of calcified nodules in the METTL3-OE group than in the NC-OE group, as further confirmed by increased ALP activity and CPC quantification analysis (Fig. 4h–k). To thoroughly examine the role of METTL3-mediated m6A modification in the process of bone regeneration, we generated a bilateral mandibular defect model in SD rats. As expected, compared with the control group, the METTL3-overexpressing SCAP group exhibited more bone-like structures, as shown by micro-CT and H&E staining (Fig. 4l, m). Collectively, these findings suggest that METTL3-mediated m6A modification potentially exerts a beneficial effect on the odonto/osteogenic differentiation of SCAPs.
a Structural diagram of lentivirus-mediated infection of SCAPs with METTL3-OE or NC-OE. b, c Immunofluorescence staining and qRT‒PCR were used to verify the efficiency of METTL3-OE and NC-OE infection at 72 h in SCAPs. Scale bar = 100 µm. d m6A dot blot assay showing that METTL3 overexpression increased m6A levels in SCAPs. e qRT‒PCR showed that METTL3 overexpression increased the mRNA expression of DSPP, DMP1, ALP, RUNX2 and OSX. f, g Western blot analysis of METTL3, DSPP, DMP1, ALP, RUNX2 and OSX after METTL3 overexpression. h–k Representative images and relevant quantification of ARS-stained ALP staining and mineralized nodule formation. Scale bar = 200 µm. l, m Micro-CT and H&E staining were used to verify the role of METTL3 in regulating SCAP osteogenesis. S: scaffold; B: bone-like tissue; scale bar = 100 µm. n = 3, * indicates P < 0.05; ** indicates P < 0.01; *** indicates P < 0.001.
Pre-miR-665/miR-665 is a functional target of METTL3 in SCAPs
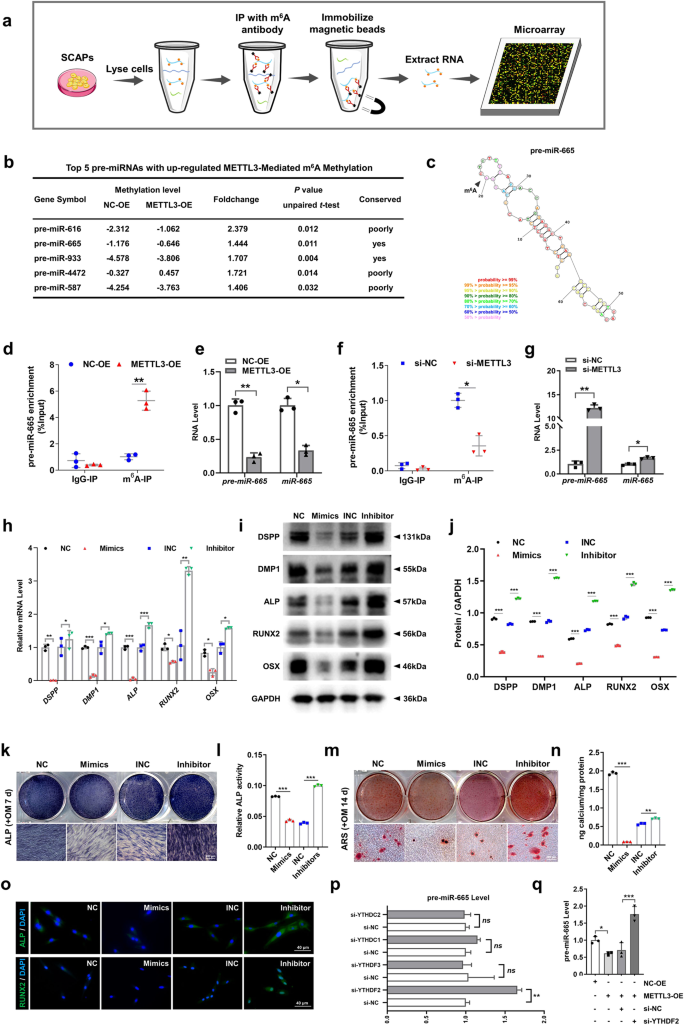
After identifying the METTL3-mediated promotion of SCAP odonto/osteogenic differentiation, we investigated the molecular mechanism through which METTL3 exerts its regulatory effects. Previous research has suggested that METTL3 can decrease the levels of mature miRNAs by modifying hairpin-shaped precursor miRNAs (pre-miRNAs) with m6A, thereby indirectly increasing the expression of miRNA target genes and ultimately participating in the regulation of cell differentiation23,24. Hence, to determine the target pre-miRNAs of METTL3, we performed a m6A RNA immunoprecipitation (RIP) microarray analysis of METTL3-OE- and NC-OE-infected SCAPs (Fig. 5a). The resulting list of the top 5 m6A-methylated pre-miRNAs in SCAPs was compiled. Among these, pre-miR-665 and pre-miR-933 exhibited the highest levels of m6A methylation ( > 1.2-fold change) and displayed a high degree of conservation across different species (Fig. 5b, c). Notably, miR-665 was found to promote the transition from acetylation to methylation of the promoters of the odontogenic differentiation markers DSPP and DMP125, suggesting the potential role of this molecule as a repressor during odontoblast induction. Consequently, we screened pre-miR-665 as a promising candidate for further investigation of METTL3 activity.
a Flowchart of the m6A-RIP microarray analysis procedure used to identify the targets of METTL3. b The top 5 pre-miRNAs with upregulated METTL3-mediated m6A methylation are listed. c The combined sites of pre-miR-665 and m6A methylation are labeled. d MeRIP-qPCR showed the positive effect of METTL3-OE on pre-miR-665 methylation by m6A compared with that of NC-OE. e qRT‒PCR analysis of the inhibitory impact of pre-miR-665 and miR-665 on METTL3 overexpression. f MeRIP-qPCR showed the negative effect of si-METTL3 on pre-miR-665 methylation by m6A compared with that of si-NC. g qRT‒PCR analysis of the ability of pre-miR-665 and miR-665 to promote METTL3 inhibition. h The mRNA expression of DSPP, DMP1, ALP, RUNX2 and OSX in the SCAPs transfected with miR-665 mimics and inhibitors. i, j The protein expression of DSPP, DMP1, ALP, RUNX2 and OSX and quantitative analysis in the SCAPs transfected with miR-665 mimics and inhibitors. k–n Representative images of ALP- and ARS-stained transfected SCAPs at 7 and 14 days in mineralized medium. Scale bar = 200 µm. o Immunofluorescence staining results of transfected SCAPs stained for ALP and RUNX2. Scale bar = 40 µm. p qRT‒PCR indicated pre-miR-665 was a target of YTHDF2. q si-YTHDF2 increased the expression of pre-miR-665 after cotransfection of METTL3-OE. n = 3, * indicates P < 0.05; ** indicates P < 0.01; *** indicates P < 0.001.
Subsequently, we investigated the effects of amplifying or suppressing METTL3 on alterations in the m6A levels of pre-miR-665 utilizing MeRIP-qPCR. MeRIP-qPCR revealed that increased METTL3 expression led to an increase in the m6A methylation level of pre-miR-665, while METTL3 inhibition had the opposite effect (Fig. 5d, f). Furthermore, the expression levels of both pre-miR-665 and miR-665 were notably decreased when METTL3 was overexpressed in SCAPs. In contrast, si-METTL3 increased the production of pre-miR-665 and miR-665, indicating that METTL3-induced pre-miR-665 m6A enrichment can suppress miR-665 expression (Fig. 5e, g).
To verify whether METTL3 regulates the odonto/osteogenic differentiation of SCAPs by mediating the m6A modification of pre-miR-665, we further examined the impact of miR-665 on the odonto/osteogenic differentiation of SCAPs. Transient transfection of miR-665 mimics and inhibitors was conducted (Supplementary Fig. 4a, b). qRT‒PCR and western blotting demonstrated that miR-665 mimics hindered the odonto/osteoblast differentiation of SCAPs, while miR-665 inhibitors increased the expression of relevant proteins and genes (Fig. 5h‒j). Furthermore, ALP and ARS staining, along with subsequent quantitative analyses, was performed, revealing a diminished degree of odonto/osteogenesis and mineralization in the SCAPs transfected with miR-665 mimics, while miR-665 inhibitors resulted in increased formation of calcium nodules. The immunofluorescence staining results also demonstrated an identical pattern (Fig. 5k–o).
The involvement of YTH domain family proteins, specifically YTHDF2/3 and YTHDC1/2, in m6A-related RNA degradation has been demonstrated26,27,28,29. Consequently, we investigated the presence of collaborative “reader” proteins in the process of METTL3-mediated pre-miR-665 m6A methylation in SCAPs. si-YTHDF2 increased the expression of pre-miR-665, indicating that pre-miR-665 may be a potential target of YTHDF2 (Fig. 5p; Supplementary Fig. 4c). Furthermore, the inhibitory effect of METTL3-OE on pre-miR-665 expression was counteracted by cotransfection of SCAPs with si-YTHDF2 (Fig. 5q).
DLX3 is directly targeted by miR-665 in SCAPs
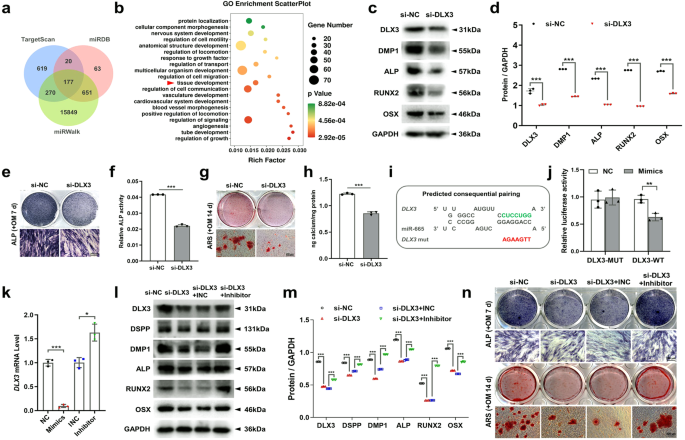
To explore the possible downstream target genes associated with the control of METTL3/miR-665 axis-mediated differentiation, we conducted a computational prediction using the TargetScan, MiRDB, and miRWalk databases to identify candidate target genes. Our analysis revealed a total of 177 potential target genes that bind to miR-665 (Fig. 6a). Moreover, Gene Ontology (GO) analysis indicated a significant correlation between these target genes and diverse cellular biological processes (Fig. 6b). These three databases predicted that DLX3 can simultaneously bind to miR-665, and GO analysis indicated that DLX3 is involved in tissue development. Furthermore, previous research has demonstrated the contribution of DLX3 to the odonto/osteoblastic differentiation of human DPSCs, and DLX3 mutations have been linked to abnormal tooth development30,31. Additionally, a significant association between DLX3 and bone mass has been reported32. Interestingly, Heair et al. revealed that miR-665 decreased DLX3 mRNA expression in dental pulp cells25. Therefore, the selection of DLX3 as a target of miR-665 is justified in SCAPs.
a Venn diagram showing the number of miR-665 target genes predicted by the TargetScan, miRDB and miRWalk algorithms. b In-depth analysis of 177 target genes through GO analysis. c, d The protein expression of DMP1, ALP, RUNX2 and OSX in the SCAPs transfected with si-DLX3. e–h Representative images of ALP staining and ARS staining in the SCAPs transfected with si-DLX3 indicating decreased osteogenic differentiation. Scale bar = 200 µm. i, j Luciferase reporter assays in 293 T cells indicated that miR-665 could bind to DLX3. k qRT‒PCR analysis of the expression of DLX3 in the SCAPs transfected with miR-665 mimics or inhibitors. l, m The inhibition of odonto/osteogenic protein expression caused by si-DLX3 was reversed by cotransfection of SCAPs with miR-665. n ARS staining and ALP staining demonstrated that the decrease in calcium nodules induced by si-DLX3 was abrogated by cotransfection with miR-665. Scale bar = 200 µm. n = 3, * indicates P < 0.05; ** indicates P < 0.01; *** indicates P < 0.001.
Next, SCAPs were transfected with si-DLX3 (Supplementary Fig. 5a), and our results showed that suppressing DLX3 led to a significant reduction in the odonto/osteoblastic differentiation of SCAPs (Fig. 6c–h). Additionally, a luciferase reporter assay was conducted by introducing wild-type or mutant 3’-UTR sequences into the DLX3 plasmid reporter in 293 T cells. The results revealed that cotransfection of miR-665 with the wild-type DLX3 reporter significantly reduced luciferase activity, while there was no notable alteration when the mutant-type plasmid reporter was utilized (Fig. 6i, j). Intriguingly, the overexpression of miR-665 in SCAPs resulted in a notable reduction in both the DLX3 gene and protein levels compared to those in the cells transfected with the negative control (NC). Conversely, the inhibition of miR-665 had the opposite effect (Fig. 6k; Supplementary Fig. 5b, c). Moreover, the expression of the DLX3 protein decreased upon YTHDF2 knockdown (Supplementary Fig. 5e). This observation was supported by the results obtained from western blotting assays, where the negative impact induced by si-DLX3 was nullified by miR-665 inhibitors (Fig. 6l, m). Furthermore, the restorative effect was convincingly confirmed through ALP staining and ARS staining (Fig. 6n).
METTL3-mediated regulation of DLX3 mRNA stability depends on m6A methyltransferase activity
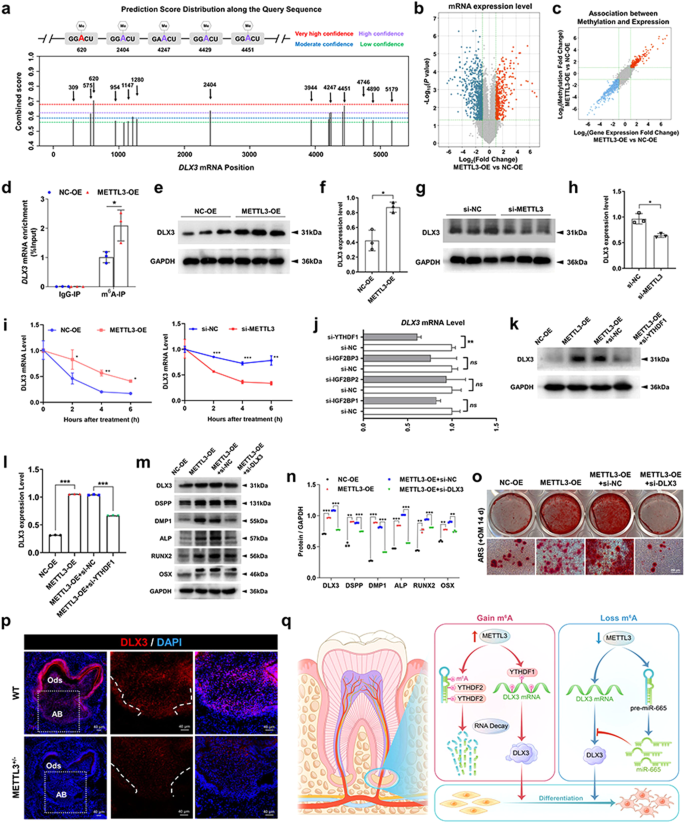
Notably, the DLX3 gene possesses 18 potential m6A modification sites, as predicted by a sequence-based m6A modification site predictor (Fig. 7a). Additionally, the findings from the m6A-RIP microarray analysis demonstrated that the METTL3-OE group exhibited elevated levels of m6A methylation and DLX3 expression compared to the NC-OE group (Fig. 7b, c). To ascertain the direct regulation of DLX3 m6A methylation by METTL3 in SCAPs, we used MeRIP-qPCR. We identified a significant increase in m6A enrichment in DLX3 upon METTL3 overexpression in the m6A-RIP group, while no change was observed in the IgG-RIP group (Fig. 7d). Moreover, the excessive expression of METTL3 led to an increase in DLX3 protein levels, while the suppression of METTL3 using si-METTL3 resulted in a reduction in the protein level of DLX3 (Fig. 7e-h). Furthermore, immunofluorescence staining confirmed the decrease in DLX3 expression in the METTL3+/− mice compared to that in their WT littermates (Fig. 7p). To further investigate whether METTL3 regulates DLX3 expression by influencing its mRNA stability, we treated SCAPs with the transcriptional inhibitor actinomycin D. RNA stability curve analysis demonstrated that METTL3-OE increased the stability of DLX3 mRNA, while si-METTL3 decreased the half-life of DLX3 mRNA (Fig. 7i).
a Potential sites and regions for m6A modification in the sequence of the DLX3 gene. b, c Screening for target genes with increased expression and m6A methylation levels in SCAPs after overexpression of METTL3 through m6A-RIP microarray sequencing. d MeRIP-qPCR illustrated that METTL3 overexpression promoted the m6A modification of the DLX3 gene in SCAPs. e–h Western blot analysis confirmed a positive correlation between METTL3 and DLX3 expression. i The half-life (t1/2) of DLX3 mRNA in the SCAPs in the control, METTL3-OE and si-METTL3 groups was measured by qRT‒PCR. j The expression of DLX3 in the SCAPs transfected with control, si-YTHDF1, si-IGF2BP1, si-IGF2BP2 and si-IGF2BP3 was verified by qRT‒PCR. k, l Western blotting results showing that the increase in DLX3 expression caused by METTL3-OE was reversed by si-YTHDF1. m, n si-DLX3 downregulated the expression of odonto/osteogenic protein markers in SCAPs in the presence of METTL3-OE. o ARS staining results illustrated that si-DLX3 slightly reversed the generation of mineralized nodules increased by METTL3-OE in SCAPs. Scale bar = 100 µm. p Immunofluorescence staining showed that compared with WT mice, METTL3+/− mice exhibited weaker DLX3 expression. Scale bar = 40 µm. q A scheme showing the role of the METTL3/pre-miR-665/DLX3 regulatory network in SCAPs (By Figdraw). n = 3, ns indicates no significance; * indicates P < 0.05; ** indicates P < 0.01; *** indicates P < 0.001.
Given the established role of YTHDF1 and IGF2BPs in promoting the stability of methylated mRNA, qRT‒PCR was used to identify the most effective “reader” protein18,33. Based on the observed inhibition of DLX3 mRNA in the SCAPs transfected with si-YTHDF1 siRNA, we hypothesized that the DLX3 transcripts are targeted by YTHDF1 (Fig. 7j; Supplementary Fig. 5d). Furthermore, the protein expression of DLX3 in SCAPs was notably reduced by si-YTHDF1 in the presence of METTL3 overexpression (Fig. 7k, l). Moreover, the upregulation of DLX3 and differentiation-associated genes induced by METTL3 overexpression was effectively counteracted by simultaneous treatment with si-DLX3 (Fig. 7m, n). ARS staining revealed that si-DLX3 decreased the presence of calcium nodules in the SCAPs of the METTL3-OE group (Fig. 7o). Based on these findings, we can infer that DLX3 is a functionally regulated target of METTL3, and its effects are dependent on YTHDF1.
As depicted in the Graphical Abstract, the odonto/osteogenic differentiation capabilities of SCAPs were significantly augmented as a result of the upregulation of METTL3. Mechanistically, METTL3 facilitated the m6A modification of pre-miR-665, which was subsequently recognized by YTHDF2, leading to its degradation and consequent suppression of miR-665 expression. As a result of this inhibition, the target gene DLX3 was ultimately expressed more strongly, thus facilitating the differentiation of SCAPs into odonto/osteogenic cells. Additionally, METTL3 directly mediates the m6A modification of DLX3. Methylated DLX3 was subsequently recognized by another m6A “reader”, YTHDF1, to ensure its stability and expression (Fig. 7q).
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- PlatoHealth. Biotech and Clinical Trials Intelligence. Access Here.
- Source: https://www.nature.com/articles/s12276-024-01245-8