Ethics declaration on animal use
C57BL/6 (B6) il33−/− mice were obtained from S. Nakae (University of Tokyo, Tokyo, Japan)38. B6-argyfp–il33−/− mice were generated by 6 times backcrossing of B6.129S4-arg1tm1lky/J (B6-arg1yfp; The Jackson Laboratory) mice on to the B6 il33−/− background at the University of Pittsburgh. St2−/− mice were originally generated on a BALB/c background39 and then backcrossed to a B6 background before use. All mice were raised in accordance with and with the express approval from the University of Pittsburgh Institutional Animal Care and Use Committee. The B6 St2fl/fl mice were provided by Giorgio Trinchieri (National Cancer Institute, Bethesda, Maryland) and crossed to B6.129P2-Lyz2tm1(cre)Ifo/J (The Jackson Laboratory) to generate LysMCre × St2fl/fl mice. Colony maintenance, including bedding and cage changes, was provided by the University of Pittsburgh Division of Laboratory Animals in Research on a weekly basis. Litters were weaned at 21 days of age and genotyping of all breeding pairs was performed to maintain the integrity of the strains. Animals were used as experimental subjects or as a source of tissues and cells. Male and female animals ≥ 8 weeks of age were used to generate MBV, whereas male animals ≥ 8 weeks of age underwent surgical manipulation. Functional testing was performed using male animals. Pain management was achieved using buprenorphine hydrochloride twice daily for 3 days and infection prevention was maintained for 3 days using Baytril. Animal weights were not recorded for the duration of the study as it was assumed that any weight change that occurred over a two-week span would be negligible. Animal weights were recorded at the time of euthanasia for animals undergoing functional testing. Euthanasia was performed in accordance with guidelines using 20–30% v/v inhaled CO2 for 8–10 min until signs of life were no longer detectable and death was ensured using cervical dislocation.
Cardiotoxin injury model
Cardiotoxin muscle injury model was used as previously described40. Briefly, a small incision was made in the skin overlying the tibias of anesthetized mice in the supine position. Blunt dissection of the adjacent subcutaneous region was used to expose the tibialis anterior (TA) muscles and 25 μl of 10 µM cardiotoxin (CTX) (Naja Pallida; Sigma Aldrich, St. Louis, MO) was administered by intramuscular injection to the left and right hindlimbs. The incision was subsequently closed using resorbable sutures and the animals allowed to recover.
Immune Cell Analysis by Flow Cytometry
Leukocytes within skeletal muscle were isolated by mincing tissue and homogenizing with gentleMACS C tubes (Miltenyi Biotec) in RPMI containing 350 U/mL type IV collagenase (Gibco) and 20 μg/mL DNase I (MilliporeSigma). Tissue was incubated in a shaker at 37 °C at speed 250 RPM for 50 min. Single-cell suspensions were then obtained by passing through a 70-μm cell strainer and purified by density gradient centrifugation using Lympholyte-M (Cedarlane). Cells were incubated with heat-inactivated goat-serum (5%) to block FcR, stained with eFluor 506L/D viability dye (Thermo Fisher) and leukocytes within the damaged muscle tissue were isolated and incubated with heat-inactivated goat-serum (5%) to block FcR, and then labelled with different combinations of fluorochrome-conjugated antibodies (BD Bioscience, Biolegend, eBioscience or MD Biosciences) to distinguish myeloid and T cell populations (CD45(30-F11), CD45.2(104), F4/80(BM8), CD8(53–6.7), CD38(90/CD38), CD4(RM4-5), Foxp3(FJK-16S), Ly6C(AL-21), B220(RA3-6B2), iNOS(CXNF7), CD206(MR5D3), CD3(17A2), CD11b(M1/70), MHCII(2G9), Ly6G(1A8), CD11c(N418), ST2(U29-93,DIH9), EGR2(erongr2), GFP(FM264G)). Data was acquired with either an Aurora-10 (Cytek) or LSRFortessa flow cytometer (BD, Bioscience) and analyzed using FlowJo, Version 10.1 (Tree Star). Antibodies: PECF594 Rat Anti-Mouse CD45 (1:400 BD Horizon Cat: 562420; Clone: 30-F11), Pacific Blue Anti-Mouse F4/80 (1:200 BioLegend Cat: 123124; Clone: BM8), BV605 Rat Anti-Mouse CD38 (1:200 BD OptiBuild Cat: 740361; Clone: 90/CD38), BV650 Rat Anti-Mouse CD4 (1:200 BD Horizon Cat: 563232; Clone: GK1.5), PE Rat Anti-Mouse Ly6C (1:200 BD Pharmigen Cat: 560592; Clone: AL-21), PE-Cy5 Rat Anti-Mouse CD45R/B220 (1:200 BD Pharmigen Cat: 553091; Clone: RA3-6B2), Alexa Fluor 700 Rat Anti-Mouse CD3 Molecular Complex (1:200 BD Pharmigen Cat: 561388; Clone: 17A2), APC/Fire 750 Anti-Mouse/Human CD11b (1:200 BioLegend Cat: 101262; Clone: M1/70), BUV395 Anti-Mouse I-A/I-E (1:200 BD OptiBuild Cat: 743876; Clone: 2G9), BUV805 Rat Anti-Mouse Ly6G (1:200 BD OptiBuild Cat: 741994; Clone: 1A8), Brilliant Violet 510 Anti-Mouse CD11c (1:200 BioLegend Cat: 117338; Clone:N418), BUV737 Rat Anti-Mouse IL33R (ST2) (1:100 BD OptiBuild Cat: 749323; Clone: U29-93), PerCP-Cyanine5.5 FoxP3 Monoclonal Antibody (1:200 Thermo Fisher Cat: 45-5773-82; Clone: FJK-16s), PE-Cyanine7 iNOS Monoclonal Antibody (1:400 Thermo Fisher Cat: 25-5920-82; Clone: CXNFT), Alexa Fluor 647 Anti-Mouse CD206 (1:200 BioLegend Cat: 141712; Clone: 068C2), eBioscience Fixable Viability Dye eFluor 506 (1:500 Thermo Fisher Cat: 65-0866-14).
Skeletal muscle immunolabeling
Macrophage immunolabeling
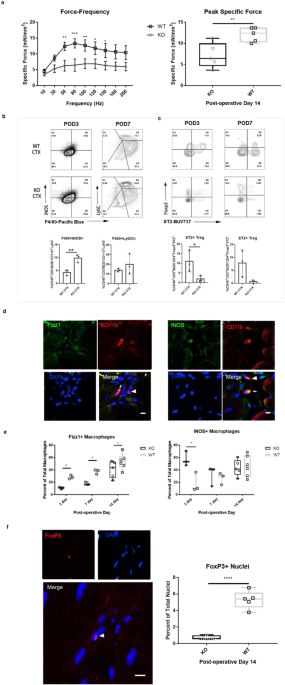
Harvesting of the tibialis anterior was performed at 3, 7, and 14-day post-injury. Each muscle was fixed in 10% neutral-buffered formalin and embedded in paraffin. 5 μm sections were cut and mounted onto glass slides. Slides were deparaffinized using xylene and ethanol gradients (100-70% EtOH). Slides were subjected to antigen retrieval and immunolabeling. Immunofluorescence was performed on serial sections for each subject and timepoint to assess the phenotypes of immune and satellite cell populations. After deparaffinization, the slides were placed in citrate antigen retrieval buffer (10 mM citric acid monohydrate, pH 6.0), microwaved at 100% power for 45 seconds, followed by 15 min at 20% power. The slides were then cooled in copper sulfate solution (10 mM CuSO4, 50 mM ammonium acetate, pH 5.0) for 20 min. Sections were then rinsed three times in Tris buffered saline/Tween 20 solution (TBST) and then incubated for 1 hour at room temperature in blocking buffer containing 0.1% Triton-X 100, 0.1% Tween, 2% goat serum, and 1% bovine serum albumin. The blocking buffer was then removed and the sections were incubated overnight at 4 °C in a humidified chamber with 1:200 rabbit-anti-CD11b (ab128797; Abcam, Cambridge, UK), a pan-macrophage marker. Following overnight incubation, each slide was washed in TBST for 3 × 2 min. A 1:200 solution of goat-anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (DAKO, Glostrup, Denmark) in blocking buffer was subsequently applied and microwaved at 40% power for 3 min in a humidified chamber and allowed to cool for 2 min before washing in TBST. After washing, sections were incubated with a 1:200 solution of red fluorescent HRP substrate (OPAL 570; Perkin Elmer, Waltham, MA) in 1x Amplification Diluent (Perkin Elmer) for 10 min and then washed in TBST. To remove anti-CD11b and anti-rabbit antibodies, sections were subjected to a second round of antigen retrieval in citrate antigen retrieval buffer, followed by cooling in copper sulfate solution, and blocked as described above. For each slide, one section was incubated with a 1:200 solution of rabbit-anti-iNOS antibody (PA-0303A; Invitrogen, Carlsbad, CA) in blocking buffer, and one section was incubated with a 1:200 solution of rabbit-anti-RELMα (500-P214; PeproTech, Rocky Hill, NJ). Slides with the primary antibodies were then placed on a raised water bath and microwaved at 40% power for 3 min, followed by 2 min of cooling. Slides were then washed in TBST solution and a 1:200 solution of goat-anti-rabbit HRP-conjugated secondary antibody was placed on the sections. Slides with secondary antibody solutions were then placed in the water bath and microwaved at 40% power for 3 min, followed by 2 min of cooling. After cooling, slides were washed in TBST and a 1:200 solution of green fluorescent HRP substrate in 1x Amplification Diluent (520 Opal, Perkin Elmer) was placed over each section and incubated in a dark humidified chamber for 10 min at room temperature. The sections were then washed in TBST, incubated with 4′,6-diamidino-2-phenylindole (DAPI) nuclear counterstain for 5 min. The sections were washed with TBST and subsequently mounted for imaging by fluorescence microscopy. For each technical replicate, the number of CD11b+ cells was counted and summed to give a total number of CD11b+ cells per biological replicate. For each biological replicate, a minimum of 2000 CD11b+ cells were counted. Co-positive macrophages, or macrophages that expressed CD11b as well as iNOS or Fizz1, the percent of total macrophages was determined by dividing the number of co-positive macrophages by the total number of CD11b+ cells.
FoxP3 staining
Paraffin embedded sections were cut and deparaffinized as described above. Antigen retrieval was performed for 20 min at 95–100 °C in citrate antigen retrieval buffer, background fluorescence was quenched using copper sulfate solution and the sections were incubated for 1 h in blocking buffer. After blocking, sections were incubated at 4 °C in a humidified chamber with either a 1:200 solution of rabbit-anti-FoxP3 (ab75763, Abcam) in blocking buffer. Following overnight incubation, slides were washed in TBST, and incubated for 1 h at room temperature with a 1:200 solution of goat-anti-rabbit HRP-conjugated secondary antibody (DAKO). Sections were then washed with TBST, nuclei were counterstained with DAPI for 5 min, and the slides were coverslipped for fluorescence microscopy. For each technical replicate, the FoxP3+ nuclei were counted and summed to give a total number of FoxP3+ nuclei per biological replicate. For each biological replicate, a minimum of 2000 nuclei were counted.
In Situ Contractile Testing
Fourteen days post-cardiotoxin injury (POD-14), in situ contractile testing protocol was implemented to evaluate the muscle’s force producing capacity41,42. Contractile testing was performed using an in situ testing apparatus (Model 809B, Aurora Scientific Inc, Canada), stimulator (Model 701C, Aurora Scientific Inc, Canada), and force transducer (Aurora Scientific Inc, Canada). Animals were anesthetized with 2% isoflurane. Through a small incision lateral to the knee, the peroneal nerve was isolated and exposed. The Achilles tendon was surgically cut using a scalpel prior to placing the animal supine on a 37 °C-heated platform. The foot was taped to the footplate with a surgical cloth tape, with the ankle position at 20° of plantarflexion (the position determined to result in the greatest force output)43. The needle electrodes were inserted beneath the skin, over the peroneal nerve. The single-twitch protocol was implemented to evaluate muscle cross-sectional area (CSA), muscle peak twitch, time to peak twitch, and half-relaxation time. Next, a force-frequency protocol was implemented by eliciting stimulations at 10, 30, 50, 80, 100, 120, 150 Hz, with a 2-minute rest between each frequency. Note that the output from the machine is torque (mN-m). Force was calculated by dividing the torque by the length of the foot plate (0.03 m). The mean CSA of the muscle was obtained using the formula: Mean CSA = (weight of TA in mg/ (length of TA in mm * density of the muscle)), where muscle density is assumed to be 1.06 g/cm344. The specific force was then obtained by dividing the force by the mean CSA. The TA muscles were subsequently harvested and fixed in formalin for histological analysis. Peak specific force was defined as the maximum force produced during the force-frequency sweep, regardless of the frequency at which it was produced.
MBV isolation and quantification
Small intestine was isolated from experimentally naïve mice B6 il33+/+ and B6 il33−/− between 6–8 weeks of age. Harvested intestines were cut into ~3 cm long segments, the tunica serosa, mucosa, and muscularis externa were removed through mechanical and chemical methods as previously described45,46,47. The resulting tunica submucosa was subsequently digested overnight at 37 °C while rotating using 0.01 mg/mL solution of Liberase DL (Sigma Aldrich, St Louis, MO) in a buffer consisting of 50 mM tris (pH 8), 5 mM CaCl2, and 200 mM NaCl. Crude digest mixtures were then subjected to progressive centrifugation: 3 × 10 min at 500 × g, 3 × 20 min at 2500 × g, and 3 × 30 min at 10000 × g. Digests were then passed through a 0.22 μm syringe filter and ultracentrifugated at 100,000 × g for 2 h at 4 °C (Beckman Coulter Optima L-90K ultracentrifuge, Brea, CA). Following ultracentrifugation, supernatants were discarded and the pellet resuspended in 1 mL of particle-free PBS and further purified via size exclusion chromatography with a 10 cm column height and 1 mL fraction volume (Sepharose CL-2B beads, Sigma). Purified MBV, contained in fractions 3–5, were collected and concentrated using 100 kDa MW cut-off spin columns (Millipore, Burlington, MA). Particle concentration of each sample was determined using a NanoSight particle counter equipped with nanoparticle tracking analysis (NTA, NanoSight, Salisbury, UK).
Exogenous MBV delivery
Two days following injury, il33−/− mice were anesthetized using 1.5–2% inhaled isoflurane and the original incision used for CTX injection was reopened. 30 μL of IL-33+ MBV were gradually delivered in 1X PBS to both hindlimb TAs intramuscularly while retracting the needle from distal to proximal tendon. Pressure was applied to prevent leakage and the skin was subsequently closed with resorbable sutures and the animals were allowed to recover under supervision on a heated surface.
Macrophage isolation, culture, and activation
Bone marrow was isolated as previously described34. Briefly, femurs, tibias, and fibulas were harvested from 6–8 week old mice and washed 3x in macrophage Complete Medium composed of 10% FBS (Invitrogen, Carlsbad, CA), 10% L929 supernatant, 10 mM non-essential amino acids (Gibco), 10 mM HEPES (Gibco), 2 mM L-glutamine (Gibco), 100 U/mL penicillin, 100 μg/mL streptomycin and 0.1% β-mercaptoethanol in DMEM high glucose (Gibco). Complete medium was flushed through the medullary space of harvested bones and plated at 2E6 cells/mL into 6 well plates (Corning) for 7 days until mature macrophages were obtained. Medium was supplemented 24 h after plating and changed every 48 hours thereafter. Mature macrophages were subsequently treated for 16 h with Complete Medium containing one of the following treatments: complete medium (M0 control), 20 ng/mL IFNγ + 100 ng/mL LPS (M1), 20 ng/mL IL-4 (M2), 20 ng/mL rIL-33, 1E9 IL-33– MBV/mL, or 1E9 IL-33+ MBV/mL. If conditioned media was collected, treatments were removed and the cells washed with PBS and 500 μL of DMEM high glucose was added for 5 h. For RNA sequencing, total RNA was isolated using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA quantity was determined using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE).
Quantitative polymerase chain reaction assays
RNA isolated from wildtype bone marrow-derived macrophages treated with 20 ng/mL rIL-33, 1 × 109 MBV/mL IL-33−, or 1 × 109 MBV/mL IL-33+ MBV were used for quantitative polymerase chain reaction (qPCR) as previously described25. Briefly, 500 ng of RNA was converted to cDNA using SuperScript III First Strand Synthesis System (Thermo Fisher) according to manufacturer’s instructions. SYBR Green (ABI) was then used to determine relative gene expression of the following transcripts: nos2, tnfa, il1b, ccl2, il6, and gapdh. Results were analyzed using the ΔΔCt method and normalized to gapdh. Primer sequences are listed in Supplementary Table 2. Fold change was calculated with respect to M0 controls for each replicate and averaged across replicates. Differences were assessed using two-way ANOVA with post-hoc testing.
Primary muscle stem cell isolation and culture
Muscle stem cells (MuSCs) were isolated from the hindlimb skeletal muscle of C57BL/6 mice. Harvested muscle tissue was washed using Wash Medium consisting of HBSS with 10% horse serum and 1% penicillin/streptomycin. Hair and tendon tissue were removed and the muscle tissue was minced until it could pass through a 10 mL serological pipette. Minced muscle tissue was then subjected to serial enzymatic digestion, beginning with 750 U/mL Collagenase II in Wash Medium (Gibco, Grand Island, NY) for 60 min at 37 °C. Collagenase digested muscle was subsequently centrifuged at 900 rpm for 5 min, and the supernatant was removed. A solution of 2.4 U/mL Dispase neutral protease (Gibco) and 750 U/mL Collagenase II in Wash Medium was then added to the cell pellet. The cells were resuspended by repeated passage through a 10 mL serological pipette, and incubated for 45 min at 37 °C. The mixture was then centrifuged at 900 rpm for 5 min, the supernatant removed, and a solution of 0.1% trypsin in Wash Medium added, and incubated at 37 °C for 30 min. After completing the final enzymatic digestion, the cell mixture was centrifuged again at 900 rpm for 5 min, the supernatant removed and the cells resuspended in Wash Medium. The cell suspension was subdivided for flow sorting. The primary sort tube was incubated for 1 h with all antibodies used for selection: CD31/CD45-FITC, Sca1-APC, and ITGA7-PeCy7. A second control tube was incubated with CD31/CD45-FITC alone, and a third tube was incubated with Propidium Iodide. Compensation beads (Ultracomp eBeads, ThermoFisher Scientific, Waltham, MA) were incubated for 30 min with individual antibodies and were used to establish gating parameters. MuSCs were isolated at ≥ 95% purity by selecting for the CD31−/45−/Sca1−/ITGA7+ population43,48.
MuSC differentiation assay
MuSC were seeded at 35,000 cells/mL into 4-well coverslipped chamber slides (Nunc Lab-Tek II chambered coverglass, Thermo Fisher) in MuSC Proliferation Medium containing 10% FBS (Invitrogen, Carlsbad, CA), 10% Horse Serum (Gibco), 100 U/mL penicillin, and 100 μg/mL streptomycin in DMEM high glucose (Gibco). Upon reaching ~80–90% confluency, MuSC proliferation medium was removed and replaced with low serum MuSC Differentiation Medium consisting of 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin in DMEM high glucose and one of the following treatments (direct treatment): 20 ng/mL recombinant IL-33 (rIL-33), 1 × 109 IL-33– MBV/mL, or 1 × 109 IL-33+ MBV/mL (MBV-WT). When macrophage conditioned media were used, Proliferation Media was replaced with a 1:1 mixture of 2X Differentiation Media and one of the following treatments were added: 20 ng/mL IFNγ + 100 ng/mL LPS-treated st2−/− macrophage conditioned media (M1 sup.), 20 ng/mL IL-4-treated st2−/− macrophage conditioned media (M2 sup.), 20 ng/mL rIL-33-treated st2−/− macrophage conditioned media (rIL-33 sup.), 1 × 109 IL-33– MBV/mL-treated macrophage conditioned media (KO MBV sup.), or 1 × 109 IL-33+ MBV/mL-treated macrophage conditioned media (WT MBV sup.). Cells were allowed to differentiate for ~4 days, after which the treatment was removed, the cells washed in PBS, and fixed in 4% paraformaldehyde (PFA) for 20 min at room temperature. PFA was then removed, the cells washed 3x in PBS and blocked at room temperature in blocking buffer consisting of 0.1% Triton X, 0.1% Tween, 2% low IgG bovine serum albumin (BSA, Thermo Fisher Scientific), 4% normal goat serum (Thermo Fisher) in PBS. After 1 h, blocking buffer was removed and 1:400 mouse-anti-myosin heavy chain (MF-20c, DSHB, University of Iowa, IA) in blocking buffer was added overnight at 4 °C. The primary antibody was then removed and the cells washed 3x in PBS, and 1:500 AF546-conjugated rabbit-anti-mouse secondary antibody (Thermo Fisher) in blocking buffer was added in the dark. After 1 h, secondary antibody was removed, cells were washed 3x in PBS, and nuclei counterstained with 4′,6-diamidino-2-phenylindole (DAPI). Cells were imaged using the 10X or 20X objectives of inverted fluorescence microscope (Axio Observer Z1, Carl Zeiss, Germany). Images were quantified using CellProfiler and the fusion index was calculated as the number of nuclei contained within myotubes with ≥ 3 nuclei/tube divided by the total number of nuclei.
Histology
Tibialis anterior muscle of wildtype mice was isolated 6–8 days post-cardiotoxin injury and MBV treatment. Tissues were fixed in formalin, paraffin-embedded, sectioned (4 µm) and stained with Masson’s trichrome using standard protocols. Using QuPath: Open source software for digital pathology image analysis49, blue fibrosis areas were quantified using the Train Object Classifier function. Trichrome percentage was quantified using blue fibrosis areas (mm2) divided by total area (mm2).
Statistical analysis
Dependent variables were assessed using one-way or repeated measures ANOVA, independent samples t-tests. Tukey’s HSD or Sidak post-hoc testing was performed for one-way and repeated measures ANOVA, respectively. Means comparisons were performed when appropriate with an applied alpha of 0.05. Data are presented as means ± SEM unless otherwise specified. Statistical testing was performed using Graphpad Prism 8 (Graphpad, La Jolla, CA).
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- PlatoHealth. Biotech and Clinical Trials Intelligence. Access Here.
- Source: https://www.nature.com/articles/s41536-024-00346-2
