Ethics statement
This project has been approved by the MWG Research Panel under the project code MWG230104. No animals were harmed in the preparation of this manuscript.
Resource availability
This study generated new unique resources. Further information and requests for resources and reagents should be directed to the corresponding author.
Access to skin samples
The IMCB-ESCAR laboratory is part of the Mandai Wildlife Group, which is the steward of Mandai Wildlife Reserve, home to Singapore Zoo, Night Safari, River Wonders, and Bird Paradise. Animals either died of natural causes or were humanely euthanised due to medical reasons. Skin samples were obtained from only one donor animal per species.
Derivation and culture of NHP primary fibroblasts
At post-mortem, the animals’ skin was cleaned with 70% ethanol and shaved before aseptic surgical preparation of the area of interest. A sterile scalpel blade was used to harvest a 3 cm x 3 cm sample of full-thickness skin. In sterile conditions, any remaining fur, fat, and epidermis were removed from the skin sample, and the sample was cut into smaller pieces. After washing in PBS, the pieces were incubated in DMEM, 1X Antibiotic–Antimycotic, and 0.5 mg/ml Fungin™, at room temperature for 30 min. After further washing in PBS, the pieces were placed in a 0.1% gelatin-coated sterile tissue culture dishes with media containing DMEM, 20% FBS, 1X MEM non-essential amino acids (NEAA), 1X Penicillin–Streptomycin, and 2 mM L-glutamine, and incubated at 37 degrees Celsius and 5% CO2. When confluent, fibroblasts were passaged with 0.25% trypsin and maintained on 0.1% gelatin-coated sterile tissue culture dishes with fibroblast media (FM) containing DMEM, 10% FBS, 1X MEM NEAA, 100 µM β-mercaptoethanol, 1X Penicillin–Streptomycin, and 2 mM L-glutamine. For efficient reprogramming, fibroblasts were transduced or transfected at passage 5 or earlier.
Derivation and culture of Crested Macaque iPSCs
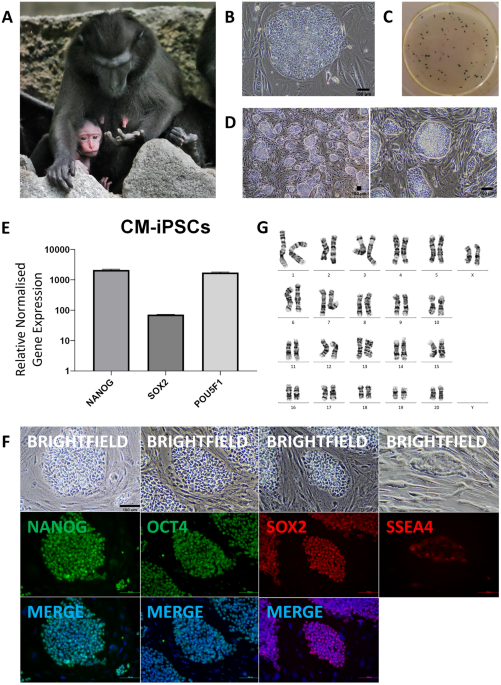
250 k Crested Macaque fibroblasts were seeded onto 0.1% gelatin-coated 3 cm dishes and cultured in FM. 24 h later (Day 0), fibroblasts were transduced with Sendai virus from the CytoTune™-iPS 2.0 kit (Thermo Fisher Scientific, A16517) containing human KLF4, OCT3/4, SOX2, and c-MYC, in 1 ml of FM. On Day 1, the media was changed to FM containing 10 ng/ml bFGF and 0.4 mM sodium butyrate, and was refreshed daily. On Day 6, cells were passaged and split 1:4 into 6-well plates coated with irradiated CF1 mouse embryonic fibroblast (iMEF) feeder layers or Matrigel®, and cultured in FM with bFGF. On Day 7, the media was changed to homemade embryonic stem cell (hES) or mTeSRTM1 media, and refreshed daily. hES media contained DMEM, 15% ESC-screened FBS, 1X MEM NEAA, 100 µM β-mercaptoethanol, 1X Penicillin–Streptomycin, 2 mM L-glutamine, 10 ng/ml bFGF, and 10 ng/ml hLIF. mTeSRTM1 media contained 0.5 mM sodium butyrate until Day 11. Reprogramming efficiency was calculated at Day 20, and was derived from dividing the resultant number of colonies as visualised on alkaline phosphatase (AP) staining by the starting cell number and an estimated transduction efficiency of 90%. iPSC colonies were picked from Day 20, and expanded and subsequently maintained in 6-well plates seeded with feeders in hES media. iPSCs were stable and could be passaged for more than 20 passages. Seven iPSC lines were generated and one was used for further analysis.
Derivation and culture of Lar Gibbon iPSCs
250 k Lar Gibbon fibroblasts were seeded onto 0.1% gelatin-coated 3 cm dishes and cultured in FM. 24 h later (Day 0), fibroblasts were transduced with Sendai virus from the CytoTune™-iPS 2.0 kit in 1 ml of FM. On Day 1, the media was changed to FM containing 10 ng/ml bFGF and 0.4 mM sodium butyrate, and was refreshed daily. On Day 6, cells were passaged and split 1:4 into 6-well plates coated with iMEF feeder layers or Matrigel®, and cultured in FM with bFGF. On Day 7, the media was changed to hES or mTeSRTMPlus (STEMCELL Technologies, #100–0276) media, and refreshed daily. mTeSRTMPlus media contained 0.5 mM sodium butyrate until Day 11. Reprogramming efficiency was calculated at Day 22, and was derived from dividing the resultant number of colonies as visualised on alkaline phosphatase (AP) staining by the starting cell number and an estimated transduction efficiency of 90%. iPSC colonies were picked from Day 23, and expanded and subsequently maintained in 6-well plates seeded with feeders in mTeSRTMPlus. iPSCs were stable and could be passaged for more than 20 passages. Six iPSC lines were generated and one was used for further analysis.
Derivation and culture of Siamang iPSCs
500 k Siamang fibroblasts were transfected with 2ug each of plasmids pCXLE-hUL, pCXLE-hSK, pCXLE-OCT3/4-p53shRNA, and pCXWB-EBNA-1 (Addgene #27,080, #27,078, #27,077, and #37,624 respectively). Electroporation was performed via the Neon™ transfection system (Thermo Fisher Scientific, MPK5000, MPK1025) at 1650 V, 10 ms, and 2–3 pulses. Transfected cells were seeded onto 0.1% gelatin-coated 6 cm dishes and cultured in FM (Day 0). 24 h later (Day 1), the media was changed to FM containing 10 ng/ml bFGF and 1 mM sodium butyrate, and was refreshed daily. On Day 7, cells were passaged and split 1:3 into 6-well plates coated with Geltrex™ and cultured in either mTeSR™ Plus or StemMACS™ iPS-Brew XF (Miltenyi Biotec, 130–104-368). Reprogramming efficiency was calculated at Day 13, and was derived from dividing the resultant number of colonies as visualised on alkaline phosphatase (AP) staining by the starting cell number and a transfection efficiency of 55.69% as determined by FACS analysis of pCXLE-EGFP (Addgene #27,082) transfected cells at day 2 under identical conditions. iPSC colonies were picked from Day 14, and expanded and subsequently maintained in 6-well plates coated with Geltrex™, in iPSC media. iPSCs were stable and could be passaged for more than 20 passages.
Derivation and culture of Proboscis Monkey iPSCs
600 k Proboscis Monkey fibroblasts were transfected with 2ug each of plasmids pCXLE-hUL, pCXLE-hSK, pCXLE-OCT3/4-p53shRNA, and pCXWB-EBNA-1. Electroporation was performed via the Neon™ Transfection System at 1650 V, 10 ms, and 3 pulses. Transfected cells were seeded onto 0.1% gelatin-coated 6 cm dishes and cultured in FM without Penicillin–Streptomycin (Day 0). 24 h later (Day 1), the media was changed to FM containing 10 ng/ml bFGF. On Day 2, the media was changed to FM containing 10 ng/ml bFGF and 0.5 mM sodium butyrate, and was refreshed daily. On Day 7, cells were passaged and split 1:4 into 6-well plates coated with Matrigel® or Geltrex™, and cultured in a 1:1 ratio of FM and a variety of iPSC medias with 10 µM Y-27632. iPSC medias were either StemMACS™ iPS-Brew XF, PluriSTEM™ Human ES/iPS Cell Medium (MERCK, SCM130), or Essential 8™ Medium (Thermo Fisher Scientific, A1517001) with 0.5 mM sodium butyrate until and including Day 11. On Day 9, the media was changed to iPSC media only, and was refreshed every other day. iPSC colonies started forming by Day 13. Reprogramming efficiency was calculated at Day 20, and was derived from dividing the resultant number of colonies as visualised on alkaline phosphatase (AP) staining by the starting cell number and a transfection efficiency of 40.58% as determined by fluorescence-activated cell sorting (FACS) analysis of pCXLE-EGFP transfected cells at day 2 under identical conditions. iPSC colonies were picked from Day 21, and expanded and subsequently maintained in 6-well plates coated with Matrigel®, Geltrex™, or feeders, in iPSC media; different coatings were used to optimize for the best culture conditions. iPSCs were stable and could be passaged for more than 20 passages. Three iPSC lines were generated and one was used for further analysis.
Karyotyping
Cells were treated overnight with 2.5 mg/ml 5-bromo-2-deoxyuridine and 10ug/ml colcemid. Cells were then dissociated with 0.5% trypsin–EDTA for 10 min, collected into a centrifuge tube, and treated with 5 ml hypotonic solution containing 75 mM KCl and 0.8% sodium citrate for 20 min. Cells were then fixed with a series of 3:1 methanol and acetic acid fixative. The cell suspension was then dropped onto clean wet slides and allowed to air dry, and placed in a 90 degree Celsius oven prior to staining. G-banding and chromosome analyses were performed in accordance with standard procedures. 20 cells were analyzed per sample. Reference karyograms were taken from the Atlas of Mammalian Chromosomes32.
Embryoid body formation and differentiation into three germ layers
iPSCs were passaged with ReLeSR™ (STEMCELL Technologies, #100–0484) and split 1:3 into ultralow attachment 24-well plates. Cells were cultured in hES media without bFGF nor hLIF for 7 days. EBs formed were either maintained in suspension for another 7 days before being harvested for RNA extraction, or transferred to a 0.1% gelatin-coated plate for another 7 days before being fixed for immunocytochemical analysis.
Differentiation into cardiomyocytes
iPSCs were differentiated into cardiomyocytes using a modified protocol31. iPSCs were seeded into Matrigel®-coated wells and cultured in their respective preferred iPSC media. 2 days later, the media was changed to RPMI 1640, B27 supplement without insulin, 10 ng/ml Activin A, 10 ng/ml BMP4, and 10 ng/ml bFGF. After 3 days, the media was changed to RPMI 1640, B27 supplement without insulin, with 3 µM IWP-2. 4 days later, the differentiated cells were maintained in RPMI 1640 containing B27 supplement with insulin.
Quantitative RT-PCR
Total RNA was extracted using Monarch® Total RNA Miniprep Kit (New England BioLabs Inc., T2010) and reverse transcribed into cDNA with iScript™ Reverse Transcription Supermix (Bio-Rad, 1,708,840). qPCR was performed using KAPA SYBR® FAST qPCR Master Mix (2X) Universal (Kapa Biosystems, KK4618) with 2.5 ng cDNA with a primer concentration of 115 nM. Samples were run in duplicates and GAPDH was used as internal control. Primers are listed in Supplementary Table 1. To measure endogenous expression of pluripotency markers OCT4, SOX2, and NANOG, primers were designed to the 3’UTRs of the mRNA sequences. OCT4 refers to OCT4A. Detection of Sendai viral reprogramming vectors was performed using primers described in the CytoTune™-iPS 2.0 kit protocol.
Immunofluorescence staining
Attached cells were washed in PBS, fixed with 4% PFA for 10 min, and washed again. Blocking was performed at room temperature for 2 h; blocking buffer contained 2% BSA, 0.3% triton X-100, in PBS. Incubation with primary antibodies was performed at 4 degrees Celsius overnight, before washing with 0.2% tween-20 in PBS. Incubation with secondary antibodies and Hoechst was performed in the dark at room temperature for 1 h, before washing with PBS. Images were acquired on a fluorescence microscope. ImageJ was used to overlay images obtained from the fluorescence microscope, and to improve visualisation of the scale bars when necessary. Antibodies are listed in Supplementary Table 2.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- PlatoHealth. Biotech and Clinical Trials Intelligence. Access Here.
- Source: https://www.nature.com/articles/s41598-023-50510-9
