Animal experiment
Mus minutoides (obtained from the Pet Shop YANOHASHI) was exsanguinated via cervical dislocation under anesthesia with medetomidine, midazolam, and butorphanol and used for each experimental procedure. The animal experiments complied with the regulations and guidelines of the Yamaguchi University Experimental Animal Committee (No. 291) and the Animal Care and Use Committee of the University of Tokyo (P22-092), and their protocols were duly approved by the respective committees. The results of the study were also reported in accordance with the ARRIVE guidelines.
Fibroblast establishment
Primary fibroblasts were isolated from the tail tips of dead M. minutoides and M. musculus. The tail tip was attached to gelatin-coated plates and cultured in Dulbecco’s modified Eagle’s medium (DMEM; FUJILIM Wako) supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 µg/mL) at 37 °C with 5% CO2. The culture medium was replaced weekly, and the cells were passaged as necessary to monitor their migration.
Plasmid construction
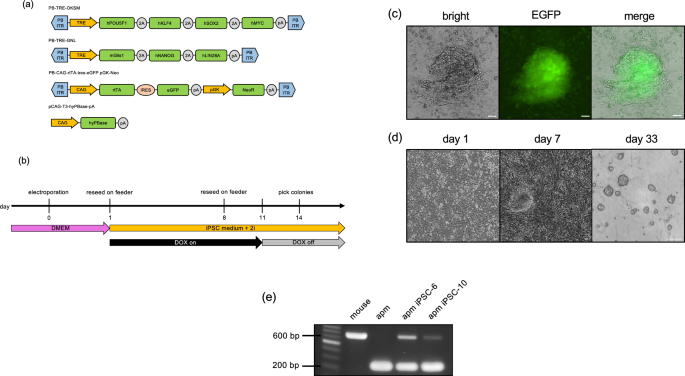
PiggyBac inverted terminal repeat (ITR) sequences were synthesized by overlap extension PCR using oligonucleotides obtained from Eurofins (Japan). The amplicons were inserted into the AatII–AflIII sites of pUC19 to construct pUC19-PB. To construct the plasmid, PB-TRE-Oct3/4-2A-Klf4-2A-Sox2-2A-cMyc (PB-TRE-OKSM), we cloned the TRE-CMV minimal promoter and a polyA signal sequence fragment from a previously constructed plasmid DNA40. The OKSM fragment was obtained from pMaster3 (Addgene plasmid #58526)41, which was modified from a previously reported plasmid42. These fragments were inserted into the ITRs of pUC19-PB.
Similarly, to construct PB-TRE-Glis1-2A-Nanog-2A-lin28a (PB-TRE-GNL), the TRE-CMV minimal promoter and poly (A) signal sequence fragments were obtained as described above. The Glis1 open reading frame (ORF) was cloned using C57BL/6-derived testis cDNA, and the NL fragment was obtained from pMaster3 (Addgene plasmid #58526)41. These fragments were inserted into the ITRs of pUC19-PB.
To construct PB-CAG-rtTA-IRES-eGFP-pGK-NeoR, the CAG-rtTA and IRES-eGFP sequences were obtained from a previously constructed plasmid DNA40. These fragments, along with pGK-NeoR (Addgene plasmid #13442)43, were inserted into pUC19-PB.
The above plasmids will be deposited in Addgene.
For pCAG-T3-hyPBase-pA construction, the hyPBase ORF was cloned from pCMV-hyPBase (kindly provided by Dr. A Bradley, Wellcome Sanger Institute) using PCR and inserted into the NotI–ClaI site of pCAG-T3-hCas9-pA (Addgene plasmid #48625)44.
The constructed vectors were sequenced using a commercial sequencing kit (Applied Biosystems, Foster City, CA, USA) and a DNA sequencer (Applied Biosystems), according to the manufacturer’s instructions. All iPSC-inducing plasmid constructs are shown in Fig. 1a.
iPSC establishment
Fibroblasts from M. minutoides were dissociated, washed, and suspended in Opti-MEM (Gibco). The cell suspension was then transferred to a 4-mm cuvette electrode (SE-204; BEX Co., LTD), and 2.5 μg/100 μL of plasmid was added individually for iPSC induction (PB-TRE-OKSM, PB-TRE-GNL, PB2-CAG-rtTA-ires-eGFP pGK-Neo, and pCAG-T3-hyPBase-pA(95)). Electroporation was performed using a CUY21EDIT II electroporator (BEX) with a single 10 ms pulse at 350 V, followed by five 50 ms pulses at 40 V at 50 ms intervals. After electroporation, the cells were reseeded on feeder cells in ESGRO Complete Basal Medium (Merck) supplemented with penicillin (100 U/mL), streptomycin (100 μg/mL), 20% KnockOut Serum Replacement (KSR) (Gibco), and ESGRO-2i Supplement Kit (Merck), with 2 μg/mL (final concentration) doxycycline (DOX) (Takara). Emerging colonies with iPSC-like morphology were passaged on feeder cells and mitomycin C-treated mouse embryonic fibroblasts and cultured with DOX until the reappearance of the colonies. Subsequently, DOX supplementation was discontinued, and the cells were further cultured. Individual colonies that maintained domed-morphology were selected under a stereomicroscope and single cells from a colony were seeded on feeder cells in 96-well plates to establish multiple M. minutoides iPSC lines.
Genomic PCR
Primers were designed based on the promoter region sequence of the M. musculus growth hormone receptor (Ghr) gene, and genomic PCR was performed using BIOTAQ DNA polymerase (NIPPON Genetics). The primer sequences are listed in Table 2. The following PCR cycling conditions were employed: initial denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 64 °C for 45 s, extension at 72 °C for 45 s, and a final extension at 72 °C for 1 min.
Alkaline phosphatase staining
Alkaline phosphatase (ALP) staining was conducted using an Alkaline Phosphatase detection kit (Merck), according to the manufacturer’s protocol.
Genomic DNA and mRNA extraction and reverse transcription
Total RNA and genomic DNA (gDNA) were extracted using the NucleoSpin TriPrep kit (Takara) or ReliaPrep RNA Cell Miniprep System (Promega), respectively, according to the respective protocols. Total RNA was reverse-transcribed using the QuantiTect Reverse Transcription Kit (QIAGEN), according to the manufacturer’s protocol.
RT-PCR
The primer design for RT-PCR was based on highly conserved regions identified by comparing the DNA sequences of M. musculus, Mus pahari, and Mus spretus. Primers were designed for the pluripotency markers: Nanog, Oct3/4, Sox2, Klf4, Eras, and Fgf4; neural markers: Tuj1 and Nestin; markers for ectoderm (Tuj1, Nestin), mesoderm (T, Acta2), cardiomyocyte (Tnnt2), endoderm (Gata4, AFP), and trophectoderm (Cdx2) differentiation in embryoid bodies (EBs) and migrating cells; and primers for the Gapdh gene. The primer sequences are listed in Table 3. PCR amplification was performed using BIOTAQ DNA polymerase (Nippon Genetics) and the following cycling conditions: initial denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 64 °C for 45 s, extension at 72 °C for 45 s, and a final extension step at 72 °C for 1 min.
Sequencing
After PCR, bands corresponding to the desired sizes were excised from the electrophoresed gel, and the amplified DNA fragments were extracted using the FastGene Gel/PCR Extraction Kit (Nippon Genetics). The extracted DNA was sequenced at Yamaguchi University Center for Gene Research. The obtained sequence data were aligned at the nucleotide and amino acid levels using CLUSTALW 2.1 software (https://www.genome.jp/tools-bin/clustalw) to determine the corresponding amino acid sequences.
RT-qPCR
Real-time PCR was conducted using the KAPA SYBR Fast qPCR Kit (Nippon Genetics) with synthesized cDNA as the template. The reaction was performed on the CFX96 Touch Deep Well real-time PCR analysis system (Bio-Rad) using the following cycling conditions: initial denaturation at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 3 s, and annealing/extension at 60 °C for 20 s. Each gene expression level was normalized to that of Gapdh and analyzed using relative cycle threshold (CT). The values are presented as mean ± standard deviation.
Cellular immunostaining
The cells were fixed with 4% paraformaldehyde for 30 min. After three washes with Tris-buffered saline (TBS), the cells were permeabilized with 0.2% Triton X-100 in TBS (TBST) for 20 min. After three additional washes with TBS, the cells were blocked with 5% goat serum in TBST for 30 min. Rabbit anti-mouse NANOG (diluted 200-fold, ab80892; Abcam) and rabbit anti-human mouse OCT4 (diluted 200-fold, C30A3C1; Cell Signaling Technology) were used as the primary antibodies for the pluripotency markers. In this study, a specific OCT4 antibody was used to recognize the pluripotency-associated OCT4A isoform rather than the more prevalent OCT4B isoform. These primary antibodies were diluted in 5% goat serum in TBST and incubated with the cells for 17 h at 4 °C. Thereafter, Alexa Fluor 594-conjugated goat anti-rabbit IgG (diluted 500-fold, ab150080; Abcam) was used as the secondary antibody. For neuronal differentiation, rat anti-mouse NESTIN antibody (diluted 200-fold, 012-26843, FUJIFILM Wako) was used as the primary antibody, and Alexa Fluor 568-conjugated goat anti-rat IgG (diluted 500-fold, A-11077; Thermo Fisher Scientific) was used as the secondary antibody. Following three additional washes with TBS, contrast staining was performed by incubating the cells with 1 g/mL DAPI in TBS for 15 min under shielded light.
RNA-seq analysis
Total RNA was extracted from M. minutoides iPSCs and fibroblasts using the Maxwell RSC RNA Cell Kit (Promega), and RNA-seq analysis was performed using the NovaSeq 6000 (Illumina) at the Yamaguchi University Center for Gene Research. The number of reads was 33.5 million for fib_1, 33.1 million for fib_2, 36.7 million for iPS-6, and 33.9 million for iPS-10. The expression levels were analyzed after TPM normalization. The RNA-seq data for mouse iPSCs and fibroblasts (GSE46104; GSM1123730, GSM1123731, GSM1123732, and GSM1123733) were obtained from NCBI Gene Expression Omnibus (GEO) database using GEO RNA-seq Experiments Interactive Navigator (GREIN) (http://www.ilincs.org/apps/grein/?gse =).
Neural differentiation
To induce the differentiation of M. minutoides iPSCs into neurons, the culture medium was replaced with the Ndiff227 medium (Takara).
Embryoid body formation
Mus minutoides iPSCs cultured on the feeder cells were detached and collected. The cells were then suspended in DMEM supplemented with 10% KSR, MEM non-essential amino acid solution (100×) (Fujifilm Wako), StemSureR 10 mmol/L 2-mercaptoethanol solution (100×) (Fujifilm Wako), penicillin (100 U/mL), and streptomycin (100 µg/mL). The supernatant containing floating iPSCs was collected and cultured in a suspension for EB formation. These EBs were subjected to adherent culture to obtain migrating cells. Total RNA was extracted from EBs, and migrating cells were derived from EBs and reverse-transcribed.
Teratoma formation
A total of 2 × 105 cells were collected, suspended in Matrigel (Corning), and subcutaneously injected into 8-week-old KSN male mice. After a 2-week incubation period, the teratomas were extracted, fixed with 3.7% paraformaldehyde, embedded in paraffin, sliced into 4 µm-thick sections, and stained with hematoxylin and eosin. Genomic DNA was extracted from teratomas using the NucleoSpin DNA FFPE XS kit (Takara), according to the manufacturer’s protocol.
Chimeric mice production
Interspecies chimeras between M. musculus and M. minutoides were generated using blastocyst complementation, according to the protocols described in Nagy et al.45, with some modifications. Briefly, ICR zygotes were obtained via in vitro fertilization46 and cultured in KSOMaa-BSA medium until the blastocyst stage. M. minutoides iPSCs were microinjected into the blastocoel of 4.0–4.5 dpc ICR blastocysts, which were then transferred into the uteri of pseudopregnant recipient ICR females on day 2.5 of the pseudopregnancy, the pups were obtained through natural birth.
Statistical analysis
Statistical analysis was performed using a t-test to determine significant differences between values. p < 0.05 was considered statistically significant.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- PlatoHealth. Biotech and Clinical Trials Intelligence. Access Here.
- Source: https://www.nature.com/articles/s41598-024-53687-9
