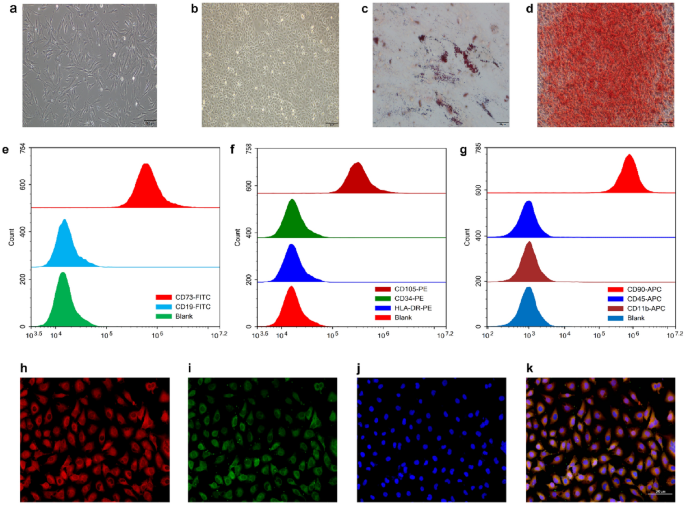
Identification and characterization of hucMSCs and HUVEC
Figure 1a,b show representative images of hucMSCs and HUVEC, respectively. Notably, hucMSCs showed fibroblast-like growth, and HUVECs showed epithelial cell-like growth. To identify the multispectral differentiation ability of hucMSCs, the differentiation of generated adipocytes and osteoblasts was identified by oil red O and alizarin red staining (Fig. 1c,d). Characterization of the surface markers of hucMSCs showed positive expression of CD73, CD90, and CD105 and negative expressions of CD11b, CD19, CD34, CD45, and HLA-DR (Fig. 1e–g). The immunofluorescence results showed that all HUVECs expressed CD31 and vWF (Fig. 1h–k). The experimental results demonstrated that both hucMSCs and HUVECs met the requirements of this experiment.
Identification and characterization of HUVECs and hucMSCs. (a) Representative pictures of hucMSCs. (b) Oil red O staining of hucMSCs for lipogenic differentiation ability. (c) Alizarin red staining of hucMSCs for osteogenic differentiation ability. (d) Representative pictures of HUVECs. (e) Flow cytometric results of CD73 and CD19, surface markers of hucMSCs. (f) Flow cytometric results for the surface markers CD34, CD105, and HLA-DR in hucMSCs. (g) Flow cytometry results of hucMSC surface markers CD11b, CD45, and CD90. (h) Immunofluorescence demonstration of HUVEC surface marker CD31 (red). (i) Immunofluorescence demonstration of HUVEC surface marker vWF (green). (j) DAPI staining (blue) of HUVEC nuclei. (k) Merged image of immunofluorescence staining of HUVECs.
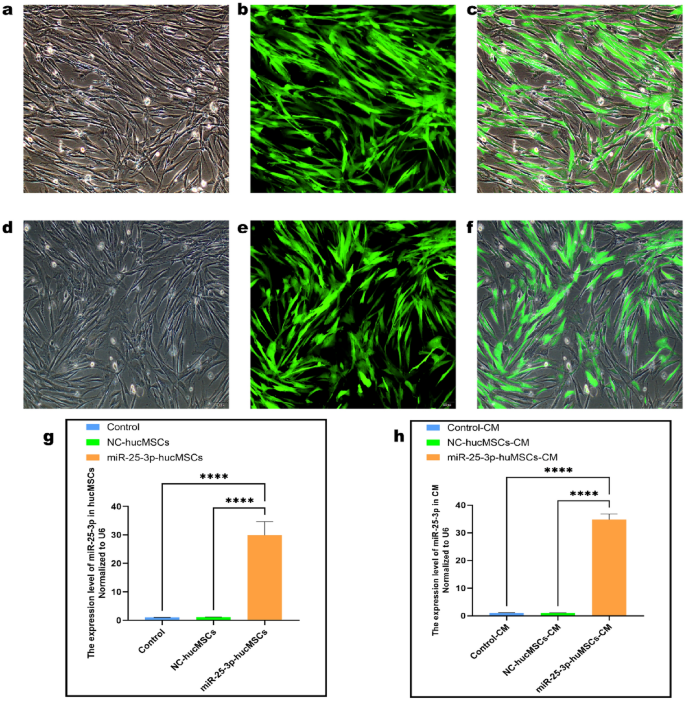
Transfection and validation of miR-25-3p lentiviral vector
We transfected hucMSCs with miR-25-3p and miR-25-3p-NC lentiviral vectors, and the multiplicity of infection (MOI) values were set to 0, 20, 50, 80, or 100. When the MOI value was adjusted to 80, nearly all of the cells were green fluorescent protein (GFP)-positive (Fig. 2a–f). Therefore, a MOI of 80 was selected as the optimal transfection value. Subsequently, a reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to validate transfection efficiency. Notably, the expression of miR-25-3p in the miR-25-3p-hucMSCs group was 28-fold (p < 0.001; Fig. 2g) higher than that in the miR-25-3p-NC-hucMSCs and hucMSCs groups. In contrast, the expression of miR-25-3p in the cell supernatants was 33-fold (p < 0.001; Fig. 2h) higher in the miR-25-3p-hucMSCs group.
Transfection and validation of miR-25-3p lentiviral vector. (a) White light plot of the miR-25-3p-hucMSC group. (b) Green fluorescence plot of the miR-25-3p-hucMSC group. (c) Merged plot of the miR-25-3p-hucMSC group. (d) White fluorescence plot of the miR-25-3p-NC-hucMSC group. (e) Green fluorescence plot of the miR-25-3p-NC-hucMSC group. (f) Merge plot of the miR-25-3p-NC-hucMSC group. (g) RT-qPCR results of miR-25-3p transfection efficiency. (h) RT-qPCR expression results of miR-25-3p in the CM of cells in the control, miR-25-3p-NC-hucMSC, and miR-25-3p-hucMSC groups.
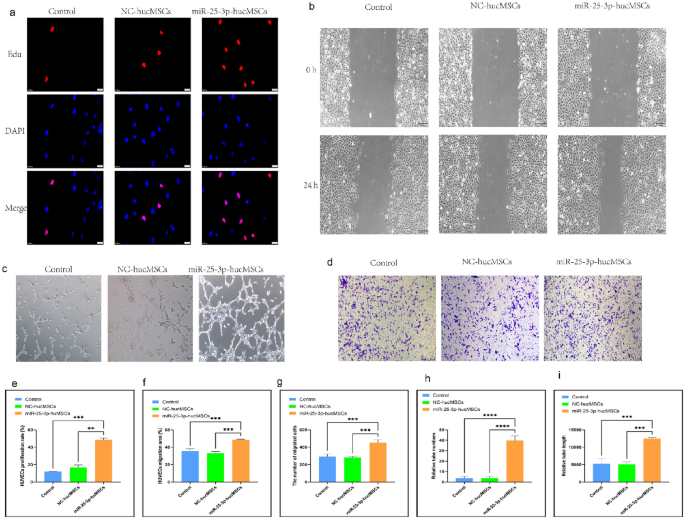
Co-culture of miR-25-3p-modified hucMSCs and HUVECs
To investigate whether hucMSCs overexpression of miR-25-3p affects the function of HUVECs, cell co-cultures were used to probe the effects of miR-25-3p on the proliferation, migration and tubule-forming ability of HUVECs.
First, the proliferative ability of HUVECs after co-culture was examined using an Edu proliferation kit. The EdU proliferation assay findings demonstrated that the positive rate of proliferating HUVECs in the miR-25-3p-hucMSC group was significantly higher than that in the control and miR-25-3p-NC-hucMSC groups, whereas that in the control and miR-25-3p-NC-hucMSCs groups was not statistically different (p < 0.001, Fig. 3a,e).
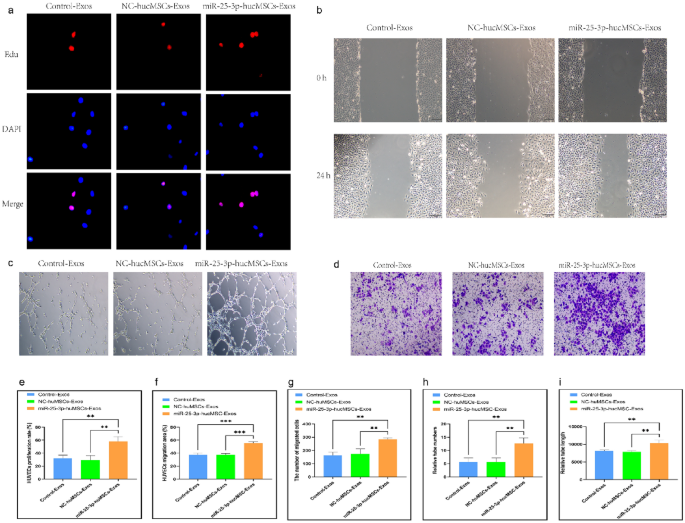
Co-culture with miRNA-25-3p-modified hucMSCs promotes HUVEC proliferation, migration, and tubule formation. (a) Representative images of the EdU proliferation experiment of HUVECs after co-culture. (b) Images of wound healing experiment of HUVECs after co-culture. (c) Images of tubule formation experiment of HUVECs after co-culture. (d) Images of transwell experiment of HUVECs after co-culture. (e) Quantification of EdU proliferation experiment of HUVECs. (f) Quantification of the area of the migrated region of HUVECs. (g) Quantification of the number of migrated HUVEC cells using the transwell assay. Quantification of the number of tubules (h) and tubule length (i) in HUVECs using a tube formation assay.
Second, scratch and transwell experiments were performed to verify the migratory capacity of HUVECs after co-culture. The scratch assay showed that miR-25-3p-modified hucMSCs significantly increased the migratory area of HUVECs (Fig. 3b,f). Similarly, the transwell assay showed that the number of HUVECs on the bottom side of the transwell chambers significantly increased in the miR-25-3p-hucMSC group (Fig. 3d,g).
Finally, tubule formation experiments were performed to verify the angiogenic capacity of HUVECs after co-culture. Notably, the number and length of formed tubules in HUVECs were significantly increased in the miR-25-3p-NC-hucMSC group compared with those in the control and miR-25-3p-NC-hucMSC groups (Fig. 3c,h,i). Therefore, miRNA126-3p-modified hucMSCs enhanced the proliferation, migration, and angiogenesis of co-cultured HUVECs through the paracrine pathway.
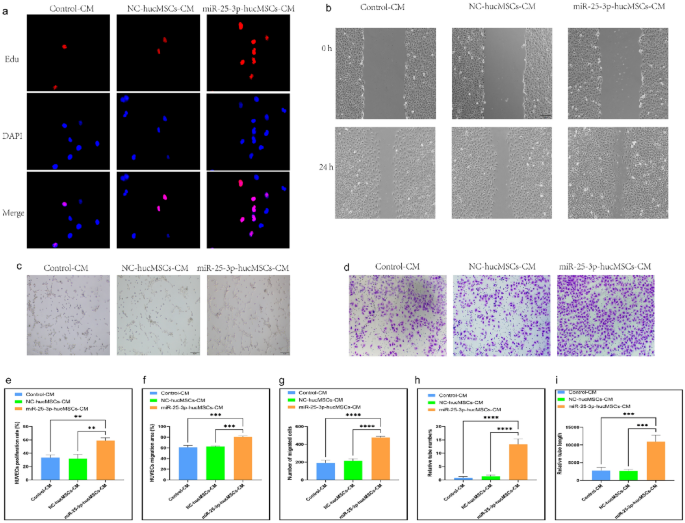
Conditioned medium (CM) from miR-25-3p-modified hucMSCs used for intervention in HUVECs
We treated HUVECs with CM to confirm whether miR-25-3p-modified hucMSCs affected the proliferation, migration, and tubule-forming abilities of HUVECs through the paracrine pathway.
The EdU proliferation assay was used to examine the proliferative capacity of HUVECs after CM intervention. Notably, the positive rate of proliferating HUVECs significantly increased with the CM intervention in the miR-25-3p-hucMSC group than in the control and miR-25-3p-NC-hucMSC groups (Fig. 4a,e).
miRNA-25-3p-modified hucMSCs-derived CM promotes HUVEC proliferation, migration, and tubule formation. (a) Representative images of the EdU proliferation assay of HUVECs. (b) Images of wound healing assay of HUVECs. (c) Images of tubule formation assay of HUVECs. (d) Images of transwell assay of HUVECs. (e) Quantification of EdU proliferation experiment of HUVECs. (f) Quantification of the area of the migrated region of HUVECs. (g) Quantification of the number of migrated HUVEC cells using the transwell assay. Quantification of the number of tubules (h) and tubule length (i) in HUVECs using a tube formation assay.
Subsequently, scratch and transwell assays were performed to verify the migration ability of HUVECs after CM intervention. The scratch assay results showed that miR-25-3p-modified hucMSC-derived CM significantly increased the migratory area of HUVECs (Fig. 4b,f). Similarly, the results of the transwell assay showed that CM in the miR-25-3p-hucMSCs group significantly increased the number of HUVECs on the bottom surface of the transwell chamber (Fig. 4d,g).
Finally, the angiogenic capacity of HUVECs after CM intervention was verified using a tubule formation assay. Notably, the CM in the miR-25-3p-NC-hucMSC group significantly increased the number and length of tubules formed by HUVECs compared with the CM in the control and miR-25-3p-NC-hucMSC groups (Fig. 4c,h,i). Therefore, miRNA126-3p-modified mucMSCs could promote the proliferation, migration, and angiogenesis of co-cultured HUVECs through the paracrine pathway.
Effect of miRNA-25-3p-modified hucMSC-derived exos intervention on HUVEC
We observed that hucMSCs influence the function of HUVEC through a paracrine pathway. Exos can affect the functions of other cells by transporting miRNAs. We intervened in HUVECs with exosomes from CM to investigate whether miR-25-3p affects the proliferation, migration, and angiogenic capacity of HUVEC through the exosomal pathway.
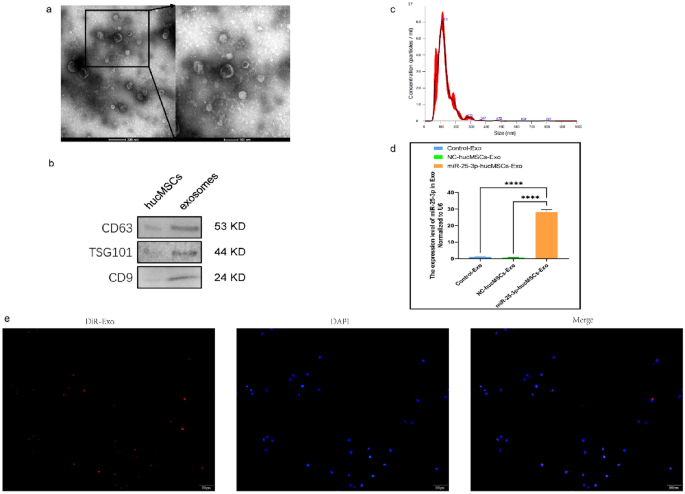
First, we extracted the exos and characterized their morphology using transmission electron microscopy (Fig. 5a) and the surface markers CD9, CD63, and TSG101 using western blotting (Fig. 5b). Nanoparticle tracking analysis (NTA) was used to determine the diameter of the exos (Fig. 5c). RT-qPCR was used to verify the differential expression of miR-25-3p in exos. Notably, miR-25-3p expression was 27-fold greater in exos produced from miR-25-3p-hucMSCs than in exos obtained from the other two groups. (Fig. 5d). Subsequently, in vitro tracking assays were performed to determine whether HUVECs could take up Dil-labeled exos. Notably, the exos were successfully internalized by HUVECs (Fig. 5e).
Exosome characterization and identification. (a) Morphology of exosomes. (b) Western blot results of markers on the surface of exosomes. (c) Results of NTA analysis of exosomes. (d) Expression of miRNA-25-3p in exosomes. (e) DiR-labeled exosomes derived from hucMSCs (red) can be internalized by DAPI-labeled HUVECs (blue).
To investigate the effects of exos derived from miR-25-3p-modified hucMSCs on the proliferation, migration, and tubule-forming ability of HUVECs, HUVECs were given 50 μg/ml of exos derived from the control, miR-25-3p-NC-hucMSC, and miRNA-25-3p-hucMSC groups. Compared with the control and miR-25-3p-NC-hucMSC groups, exos derived from the miR-25-3p-hucMSC group significantly increased proliferation (Fig. 6a,e), migration capacity (Fig. 6b,f,d,g), and in vitro angiogenic capacity (Fig. 6c,h,i) of HUVECs. Therefore, miR-25-3p-modified hucMSCs can affect HUVEC function through the exosomal delivery of miR-25-3p.
miRNA-25-3p modified hucMSCs-derived exosomes promote HUVEC proliferation, migration, and tubule formation. (a) Representative images of the EdU proliferation assay of HUVECs. (b) Images of wound healing assay of HUVECs. (c) Images of tubule formation assay of HUVECs. (d) Images of transwell assay of HUVECs. (e) Quantification of EdU proliferation experiment of HUVECs. (f) Quantification of the area of the migrated region of HUVECs. (g) Quantification of the number of migrated HUVEC cells using the transwell assay. Quantification of the number of tubules (h) and tubule length (i) in HUVECs using a tube formation assay.
Neutral sphingomyelinase 2 inhibitor (GW4869) affects HUVEC function by decreasing secretions of exos
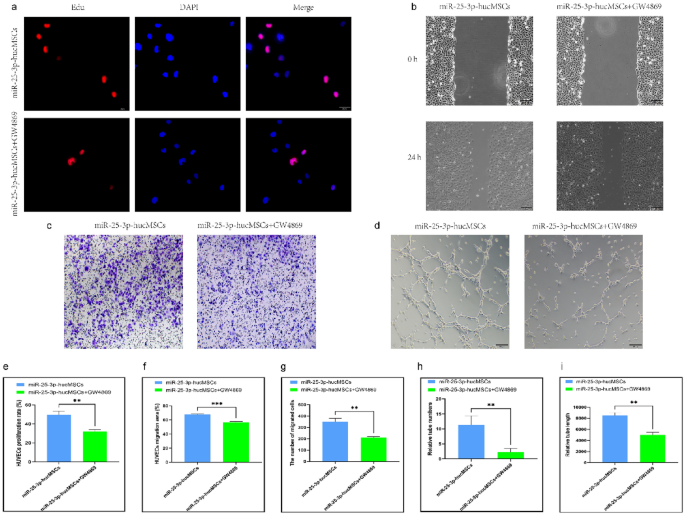
To verify whether miR-25-3p-modified hucMSCs influenced HUVEC function through exosomal secretion, we investigated the effect of exosome release inhibitors. The co-cultured HUVECs in the miR-25-3p-hucMSCs + GW4869 group showed significantly decreased proliferation (Fig. 7a,e) and migration (Fig. 7b,c,f,g) and reduced in vitro angiogenic capacity compared with the miR-25-3p-hucMSC group (Fig. 7d,h,i).
GW4869 inhibits HUVEC function after co-culture with miR-25-3p-hucMSCs. (a) Representative images of the EdU proliferation assay of HUVECs. (b) Images of wound healing assay of HUVECs. (c) Images of transwell assay of HUVECs. (d) Images of tubule formation assay of HUVECs. (e) Quantification of EdU proliferation experiment of HUVECs. (f) Quantification of the area of the migrated region of HUVECs. (g) Quantification of the number of migrated HUVEC cells using the transwell assay. Quantification of the number of tubules (h) and tubule length (i) in HUVECs using a tube formation assay.
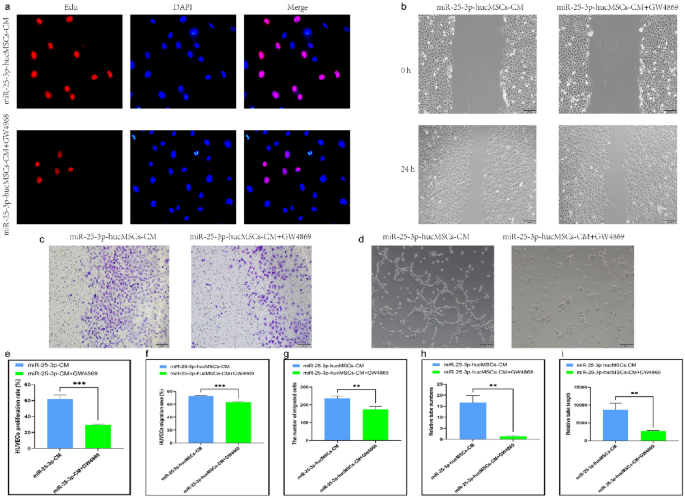
Similarly, HUVECs treated with CM in the miR-25-3p-hucMSCs + GW4869 group displayed considerably decreased proliferative activity (Fig. 8a,e) and migratory (Fig. 8b,c,f,g) and tubule-forming capacities than those in the miR-25-3p-hucMSCs group (Fig. 8d,h,i).
GW4869 inhibits function of HUVECs after intervening with miR-25-3p-hucMSCs-derived CM. (a) Representative images of the EdU proliferation assay of HUVECs. (b) Images of wound healing assay of HUVECs. (c) Images of Transwell assay of HUVECs. (d) Images of tubule formation assay of HUVECs. (e) Quantification of EdU proliferation experiment of HUVECs. (f) Quantification of the area of the migrated region of HUVECs. (g) Quantification of the number of migrated HUVEC cells using the transwell assay. Quantification of the number of tubules (h) and tubule length (i) in HUVECs using a tube formation assay.
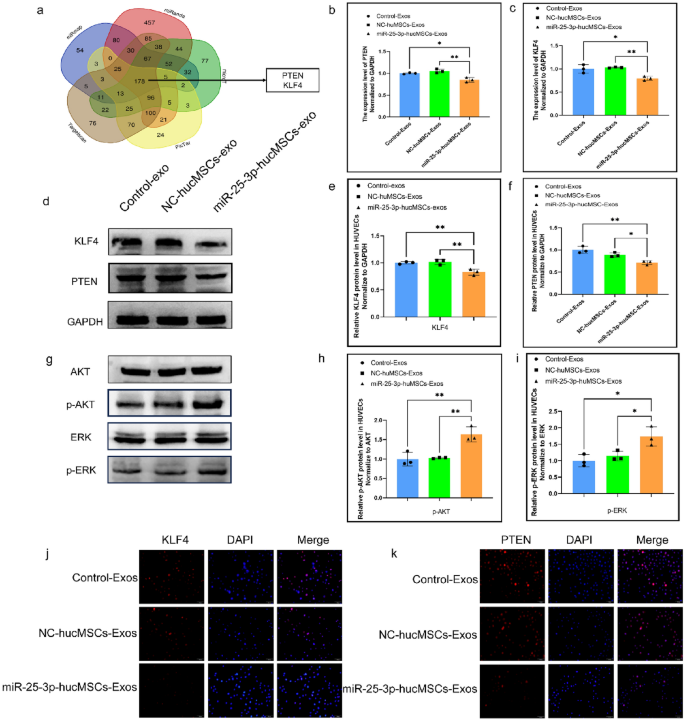
Exos derived from miR-25-3p-modified hucMSCs regulate cellular function of HUVECs by targeting KLF4 and PTEN in HUVEC
Following bioinformatics analysis and synthesis of prior studies23,24, we selected KLF4 and PTEN as probable downstream target genes of miRNA-25-3p for further research. (Fig. 9a). We extracted total proteins from HUVECs after exosome treatment for subsequent studies. The mRNA and protein expression levels of KLF4 and PTEN were significantly lower in the miRNA-25-3p-MSC group compared with the hucMSC and miRNA-25-3p-NC-hucMSC groups (Fig. 9b–f). Furthermore, the miRNA-25-3p-hucMSC group had considerably higher levels of phosphorylated AKT and ERK1/2. (Fig. 9g–i). Finally, immunofluorescence showed that PTEN and KLF4 protein levels were significantly reduced in the miR-25-3p-hucMSC-Exos group (Fig. 9j,k).
miR-25-3p-modified hucMSCs-derived exosomes regulate HUVECs function by targeting PTEN and KLF4. (a) Bioinformatic analysis of potential target genes of miR-25-3p. (b) The mRNA expression level of PTEN in HUVECs after exosomes intervention. (c) The mRNA expression level KLF4 in HUVECs after exosomes intervention. (d) The protein expression levels of PTEN and KLF4 in HUVECs after exosome intervention. (e,f) Quantification of PTEN and KLF4 protein expression levels. (g) The protein expression levels of AKT phosphorylation and ERK1/2 phosphorylation in HUVECs after exosome intervention. (h,i) Quantification of AKT phosphorylation and ERK1/2 phosphorylation protein expression levels. (j,k) Cytofluorescence verification of PTEN and KLF4 protein expression levels in HUVEC.
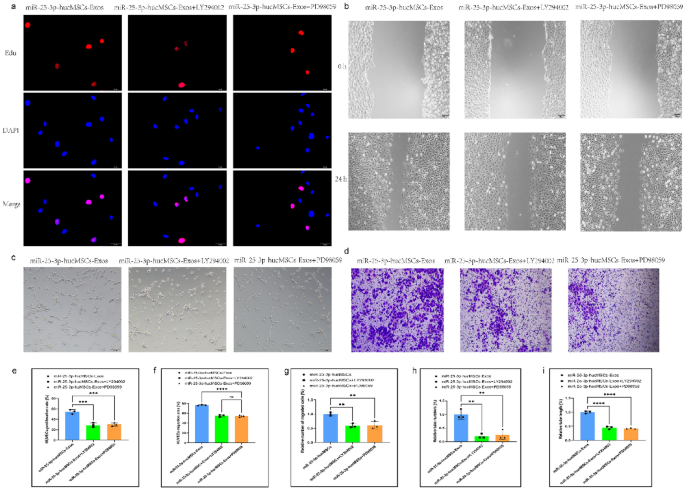
In addition, to validate the aforementioned pathways, we used the AKT inhibitor LY294002 and the ERK1/2 inhibitor PD098059 in the exosomal intervention studies. The AKT inhibitor LY294002 and the ERK1/2 inhibitor PD098059, in particular, dramatically decreased proliferation (Fig. 10a,e) and migration ability (Fig. 10b,c,f,g) of HUVECs and also reduced the number and length of tubes formed in HUVECs compared with the miR-25-3p-hucMSC group (Fig. 10d,h,i).
Signaling pathway inhibitors reversed HUVEC function in exosomal intervention experiments. (a) Representative images of the EdU proliferation assay of HUVECs. (b) Images of wound healing assay of HUVECs. (c) Images of tubule formation assay of HUVECs. (d) Images of transwell assay of HUVECs. (e) Quantification of EdU proliferation experiment of HUVECs. (f) Quantification of the area of the migrated region of HUVECs. (g) Quantification of the number of migrated HUVEC cells using the transwell assay. Quantification of the number of tubules (h) and tubule length (i) in HUVECs using a tube formation assay.
Transplantation of miR-25-3p-modified hucMSCs in the PVT model
All animal procedures were undertaken in accordance with relevant guidelines and regulations, were approved by the Ethics Committee of the First Hospital of Lanzhou University. Findings and experiments described in this paper were designed and reported following the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines.
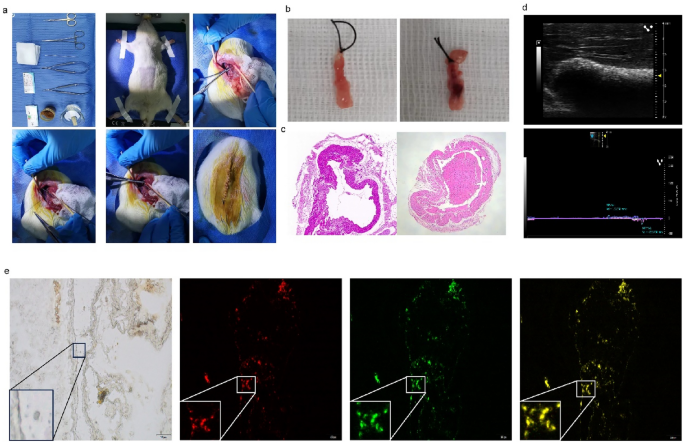
To determine whether miRNA-25-3p overexpression in hucMSCs improved portal endothelial cell repair and reduced the local inflammatory response in vivo, we used a rat PVT model for further validation. First, we constructed (Fig. 11a) and validated a rat PVT model (Fig. 11b,c). Detection of portal vein flow rate and internal diameter by small animal ultrasonography (Fig. 11d). The homing of the implanted hucMSCs was tracked via Dil labeling. The homing of the implanted hucMSCs was tracked via Dil labeling. Immunofluorescence studies showed that Dil-positive cells were predominantly located in the damaged portal vein endothelium (Fig. 11e). The damaged portal veins in the miRNA-25-3p-hucMSCs group had a higher degree of repair than those in the other groups.
Transplantation of miR-25-3p-modified hucMSCs in vivo. (a) Construction of the rat PVT model. (b) Representative picture of the portal vein under the stereomicroscope. (c) Representative pictures of the pathology of portal vein thrombosis. (d) Representative images of portal vein ultrasound. (e) Representative images of miR-25-3p-modified hucMSCs homing.
In vivo transplantation of miRNA-25-3p-modified hucMSC promotes the repair of damaged portal veins
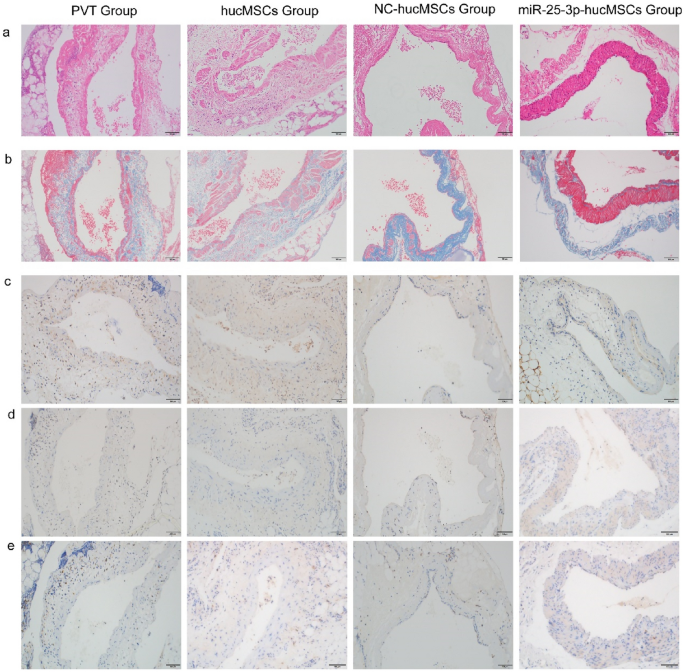
To further validate the role of miRNA-25-3p-modified hucMSCs, we transplanted miRNA-25-3p-modified hucMSCs in vivo and observed the repair of portal veins. We obtained portal veins from each group and performed hematoxylin and eosin (H&E; Fig. 12a) and Masson (Fig. 12b) staining to observe the repair of portal veins. Additionally, we performed immunohistochemical staining for Ki-67 (Fig. 12c) in the portal veins of each group to observe the proliferation of venous endothelial cells. Simultaneously, staining for TNF-α (Fig. 12d) and CD68 (Fig. 12e) was performed to observe the inflammatory factors in the portal veins and infiltration of inflammatory cells, respectively. In vivo transplantation of miRNA-25-3p-hucMSC enhanced endothelial cell proliferation while decreasing TNF-α and CD68-positive cell expression in portal veins.
In vivo transplantation of miRNA-25-3p-modified hucMSCs attenuated rat portal vein inflammation and promoted endothelial cell proliferation. (a) Representative image of the portal vein stained with H&E. (b) Representative image of the portal vein stained with Masson trichrome. (c) Representative image of the portal vein stained with TNF-α. (d) Representative image of the portal vein stained for Ki67. (e) Representative image of the portal vein stained for CD68.
Therefore, in vivo transplantation of miRNA-25-3p-hucMSC may have a favorable protective effect on damaged portal veins compared with treatment with hucMSCs or miR-25-3p-NC-hucMSCs.
- SEO Powered Content & PR Distribution. Get Amplified Today.
- PlatoData.Network Vertical Generative Ai. Empower Yourself. Access Here.
- PlatoAiStream. Web3 Intelligence. Knowledge Amplified. Access Here.
- PlatoESG. Carbon, CleanTech, Energy, Environment, Solar, Waste Management. Access Here.
- PlatoHealth. Biotech and Clinical Trials Intelligence. Access Here.
- Source: https://www.nature.com/articles/s41598-024-64263-6